Protein Folding, Misfolding and Aggregation
One of the major advantages of mass spectrometry (MS) and ion mobility-mass spectrometry (IM-MS) is the ability to measure one species in the presence of many others. This feature is already widely utilized for the analysis of complex mixtures in proteomics and metabolomics and has recently been shown to be also of exceptional use for structural biology. Instead of adopting one defined oligomerization state, some proteins exist as equilibrium of several distinctly sized species. These so-called polydisperse assemblies cannot be analyzed directly by conventional solution-phase methods, which exclusively yield data that reflect the average of the entire ensemble. With MS and IM-MS on the other hand, specific non-covalently associated oligomers can be probed directly out of their heterogeneous in vitro environment without changing the underlying equilibrium.
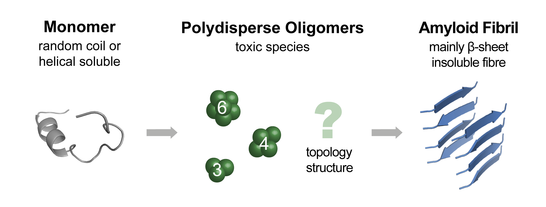
The most relevant example of polydisperse systems are probably amyloid forming proteins occurring in neurodegenerative disorders such as Alzheimer's and Parkinson's disease. These proteins undergo a spontaneous transition from a soluble, often partially folded form into an insoluble, beta-sheet rich amyloid fibril. The underlying assembly pathway is not fully understood to date, but there has been increasing evidence that oligomeric folding intermediates, which occur during the transformation process, are the toxic species in the above-mentioned diseases. Their structure however, is far from being understood. Here we aim to characterize pre-selected oligomers of polydisperse protein assemblies regarding their secondary, tertiary, and quaternary structure using a combination of MS, IM-MS and gas-phase spectroscopy techniques.
Related Publications
-
An Intrinsic Hydrophobicity Scale for Amino Acids and Its Application to Fluorinated Compounds
Hoffmann, W.; Langenhan, J.; Huhmann, S.; Moschner, R. Chang, J.; Accorsi, M.; Seo, J.; Rademann, J.; Koksch, B.; Bowers, M. T.; von Helden, G.; Pagel, K.; Angew. Chem. Intl. Ed. 2019, 58, 8216–8220. -
NFGAIL Amyloid Oligomers: The Onset of Beta-Sheet Formation and the Mechanism for Fibril Formation
Hoffmann, W.; Folmert, C.; Moschner, J.; Huang, X.; von Berlepsch, H.; Koksch, B.; Bowers, M. T.; von Helden G.; Pagel, K.; J. Am. Chem. Soc. 2018, 140, 244–249. -
Ion mobility-mass spectrometry and orthogonal gas-phase techniques to study amyloid formation and inhibition
Hoffmann, W.; von Helden, G.; Pagel, K.; Curr. Opin. Struct. Biol. 2017, 46, 7–15. -
An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies
Seo, J.; Hoffmann, W.; Warnke, S.; Huang, X.; Gewinner, S.; Schöllkopf, W.; Bowers, M. T.; von Helden, G.; and Pagel, K.; Nat. Chem. 2017, 9, 39–44.
-
Intrinsically disordered p53 and its complexes populate compact conformations in the gas phase
Pagel, K.; Natan, E.; Hall, Z.; Fersht, A.R.; Robinson, C.V.; Angew. Chem. Intl. Ed. 2013, 52, 361–365. - Following polypeptide folding and assembly with conformational switches
Pagel, K.; Koksch, B.; Curr. Opin. Chem. Biol. 2008, 12, 730–739.




