Modification of Peptides and Proteins with Fluorinated Amino Acids
One other aspect of peptide and protein research that we devote our efforts to is expanding the toolkit of amino acid building blocks to improve the pharmacokinetic and physicochemical profiles of peptide-based drug candidates and materials. Amino acid residues carrying functional groups that are orthogonal to biological systems may serve as either biophysical probes for the detailed determination of structure-activity relationships, or components of tailor-made biomolecules. In particular, the judicious incorporation of fluorine atoms has been shown to enhance protein stability in vivo and to improve the physical properties of protein-based materials (Berger, Acc. Chem. Res., 2017). We have developed peptide-based models, sensitive enough to report on changes that are brought about by the introduction of just one fluorine atom, to systematically study the complex molecular interactions of fluoroalkyl groups within native polypeptides regarding space filling, lipophilicity, and hydrogen-bonding (Jäckel, Angew. Chem. Int. Ed., 2006). We were also able to determine how fluorinated amino acids influence the structure and thermodynamic stability of the α-helical coiled coil motif (Salwiczek, ChemBioChem, 2009). Furthermore, the application of methods such as surface plasmon resonance (SPR) and phage display technology enables detailed study of the binding between natural and nonnatural peptide partners (Vagt, Org. Biomol. Chem., 2010). These studies will pave the way for optimizing the incorporation of fluorinated amino acids for the de novo design of peptide-based materials and drugs.
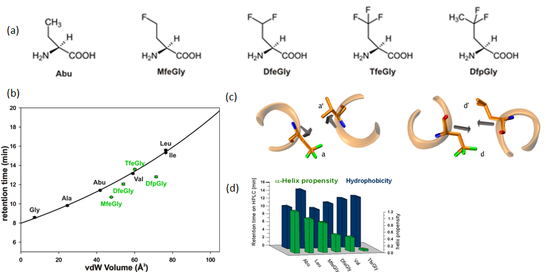
Figure 3. (a) Chemical structure of Abu-analogues with increasing fluorine content . (b) Correlation of retention time and solvent accessible surface area of amino acids of side chain fluorinated amino acids. (c) Packing of a and d positions in a parallel coiled coil dimer. (d) Bar chart depicting α-helix propensity vs. hydrophobicity.
Using a coiled coil heterodimeric model system and substitutions focused to the hydrophobic core, we have shown that the impact of size and polarity of the fluorinated building blocks highly depends upon the immediate environment of the substitution. Coiled coils are α-helical assemblies that possess a (pseudo-) repetitive primary sequence (abcdefg)n, in which hydrophobic side chains generally occupy the a- and d-positions (Figure 3). At central a positions, where the Cα-Cβ bond vectors point out of the hydrophobic core and into solution, stability is mainly determined by side chain volume. At central d positions, however, the destabilizing impact of fluorine-induced polarity prevails, since here the Cα-Cβ bond vectors point directly into the hydrophobic core (Salwiczek, Chem. Eur. J., 2009).
To efficiently screen for preferred interaction partners of the four aminobutyric acid analogues, phage display was carried out (Vagt, Org. Biomol. Chem., 2010). The predetermined secondary and tertiary structure of coiled coils are the features of our system that allow this technology to be exploited. Thus, fluorinated amino acids are incorporated into the hydrophobic positions of one strand of the heterodimer, and the residues of the other strand expected to directly interact with the nonnatural analogues are randomized to create a library that is displayed on bacteriophage. Despite the polarity that is induced by partial fluorination, the interaction profiles of all four fluoroalkylated amino acids studied so far are similar to that of aminobutyric acid. Moreover, independent of the respective microenvironment, incorporating fluorinated aminobutyric acids into a central a- or d-position of a parallel heterodimeric coiled coil leads to pairing characteristics that are similar to their canonical analogues (Nyakatura, RSC Advances, 2013).
Fluorinated amino acids have been rarely investigated in amyloid forming peptides. This is surprising since they generally show relatively low helix propensities in comparison to their native counterparts and may thus be more suitable for the incorporation into beta sheet rich structures. In order to elucidate the impact of amino acid side chain fluorination on amyloid formation, we have substituted valine residues within solvent exposed regions of a de novo designed coiled coil peptide that is able to undergo a conformational switch to beta-sheet rich amyloid fibrils. We found that these valine residues are responsible for the structural rearrangement, and resolved the internal architecture of peptide strands within the fibrils (Gerling, Biomacromol., 2011). Interestingly, the number of carbon-fluorine bonds present within the side chain has a significant effect on the kinetics of the structural transition into amyloids. Increasing the fluorine content increases the rate of folding. This effect can be explained by the interplay of several factors. Although size and hydrophobicity directly and additively increase with fluorination, α-helical propensity decreases. Thus, the incorporation of fluorinated building blocks into amyloidogenic structures could be of use in materials science applications.
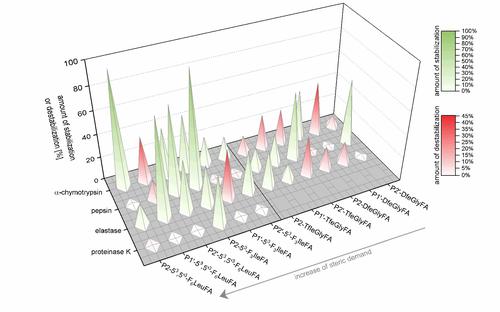
Rapid hydrolysis by proteases of the blood or digestive system usually limits the application of peptides as therapeutics. To systematically investigate the influence of fluorinated amino acids on the proteolytic stability of peptides, we synthesized a model peptide library that contained amino acids with systematically changed fluorine content at different positions relative to the protease cleavage sites. Using an analytical reversed phase HPLC assay with fluorescence detection and mass-spectrometric experiments, the proteolytic stability of the peptides was characterized.
The studies show that the impact of incorporation of side-chain fluorinated amino acids on the proteolytic stability is not predictable and cannot be generalized. Depending on the substitution position relative to the cleavage site, the chemical nature and fluorine content of the side chain, and the particular protease, fluorine substituents can alter the resistance of peptides towards proteolytic digestion in various ways. Sterically demanding, and/or highly fluorinated amino acids such as 53,5’3-F6Leu or 53-F3Ile seem to be more efficient in inducing protease resistance, but our experiments reveal that also amino acids with a smaller side chain (TfeGly and DfeGly) are able to significantly improve proteolytic stability in cases where highly fluorinated analogues clearly fail.
Our results provide valuable knowledge for the design of stable peptide-based drugs and contribute to the evolution of fluorination as a powerful tool in peptide and protein engineering.
Further Reading
S. Huhmann, B. Koksch, Fine-Tuning the Proteolytic Stability of Peptides with Fluorinated Amino Acids. Eur. J. Org. Chem. 2018, 2018, 3667-3679. https://doi.org/10.1002/ejoc.201800803
S. Huhmann, A.-K. Stegemann, K. Folmert, D. Klemczak, J. Moschner, M. Kube, B. Koksch, Position-dependent impact of hexafluoroleucine and trifluoroisoleucine on protease digestion. Beilstein J. Org. Chem. 2017, 13, 2869-2882. https://doi.org/10.3762/bjoc.13.279
V. Asante, J. Mortier, G. Wolber, B. Koksch, Impact of fluorination on proteolytic stability of peptides: a case study with α-chymotrypsin and pepsin. Amino Acids 2014, 46, 2733-2744. https://doi.org/10.1007/s00726-014-1819-7
V. Asante, J. Mortier, H. Schlüter, B. Koksch, Impact of fluorination on proteolytic stability of peptides in human blood plasma. Biorg. Med. Chem. 2013, 21, 3542-3546. https://doi.org/10.1016/j.bmc.2013.03.051
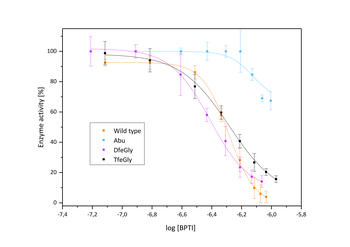
Inhibition of β-trypsin with BPTI variants
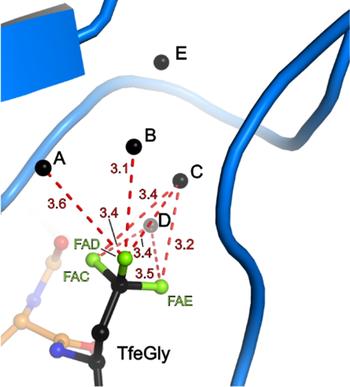
Binding pocket of β-trypsin-BPTI-TfeGly complex
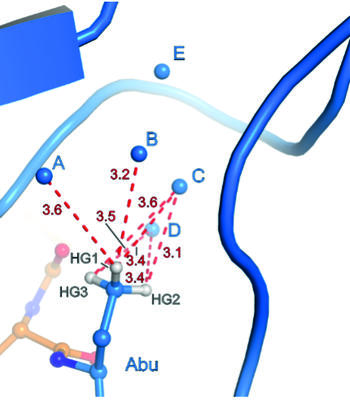
Binding pocket of β-trypsin-BPTI-Abu complex
Fluorinated amino acids have been incorporated into peptides in previous studies to investigate to what extent this unique element affects important properties, like biological activity and bioavailability. It has been shown that fluorination of the side-chains of otherwise canonical residues is in some cases advantageous, and typically does not perturb the native structure. Fewer studies have been undertaken with site-specific incorporation of fluorinated non-canonical amino acids derivatives into peptides. In general, the precise impact of fluorination is often difficult to predict and therefore requires further systematic investigation.
The presence of a single fluoroalkyl amino acid side chain (Figure, right panel) in the P1 position of the well-characterized serine-protease inhibitor BPTI (bovine pancreatic trypsin inhibitor) can fully restore inhibition properties (Figure, left panel) to a mutant containing the corresponding hydrocarbon side chain at the same position (Figure, center panel).
Further investigations revealing the impact of fluorine on protein-protein interactions are currently ongoing.
Further Reading
Ye, S.; Loll, B.; Berger, A. A.; Mülow, U.; Alings, C.; Wahl, M. C.; Koksch, B. Chem. Sci., 2015, 6, 5246-5254. (DOI:10.1039/C4SC03227F)
Understanding the influence of artificial amino acid side chains containing fluorine atoms on peptide or protein properties is undisputed important for peptide and protein engineering. For this purpose, incorporation of fluorinated amino acids (fAAs) has been applied in vitro by Solid-Phase Peptide Synthesis (SPPS), and in vivo by bacterial ribosomal translation machinery.
This study focused on the latter method, which allows the synthesis of fluorinated proteins with high fidelity, and no limitation in length. The wild type isoleucyl-tRNA synthetase (IleRS) from Escherichia coli has been used to investigate the interaction of aminoacylation and editing activities against fAAs, including small aliphatic AAs as (2S)-4,4,4-trifluoroethylglycine (TfeGly). Although IleRS activates TfeGly, we initially could not achieved the bacterial production of fluorinated peptides and proteins. The reason behind this failed attempt is the efficient editing against TfeGly, which leads to reduced dissociation rate of TfeGly-tRNAIle from the CP1 domain of IleRS and prevents the accummulation of TfeGly-tRNAIle. Taking this knowledge in to account, we used a post-transfer editing deficient mutant of IleRS, IleRSAla10, and succeeded in ribosomal incorporation of TfeGly.
By this detailed analysis of each step of ribosome-mediated translation process, we are able to provide valuable data for protein engineering towards fluorination of peptides and proteins.
Further Reading
J.-S. Völler, M. Dulic, U. I. M. Gerling-Driessen, H. Biava, T. Baumann, N. Budisa, I. Gruic-Sovulj, B. Koksch, Discovery and Investigation of Natural Editing Function against Artificial Amino Acids in Protein Translation, ACS Cent. Sci., 2017, 3 (1), 73–80.