Optofluidics at (Bio)Interfaces
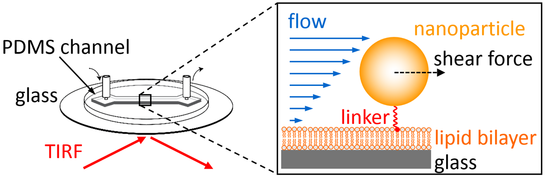
Optofluidic setups combine microfluidic channel architectures with optical microscopy, thereby allowing to characterize interfacial processes with high sensitivity and accuracy (using microscopy) in dependence of changes induced in the properties of the bulk solution (using microfluidics). We apply this concept in several projects, all of which have in common that the flow (passing through the channel) is employed to create a well-defined shear rate that, in turn, generates well-defined shear forces acting on interface-linked nanoparticles. Depending on the linking strategy used in the experiment, this shear forces will have different outcomes and hence different information can be obtained from such measurements. For example, nanoparticles can be linked using mobile linkers to a fluid-phase supported lipid bilayer (SLB), which allows the nanoparticles to move in 2D while being bound to the SLB. Application of shear forces on such mobile particles will induce a directed nanoparticle motion, which can be used to accurately quantify nanoparticle properties such as their 2D mobility, their hydrodynamic size, or their composition (flow-based mobility analyses). In contrast, nanoparticles can also be linked directly to an immobile interface. Due to use of immobilized linkers, such nanoparticles cannot move freely but will load the linkers with a (well-defined) fraction of the acting shear force, which can be used to probe nanomechanical properties of molecules (flow-based force measurements). Both strategies will be detailed below.
Flow-based mobility analyses
Recently, we proposed to analyze the 2D motion of nanoparticles, which are linked by mobile linkers to a fluid phase-SLB and which are subject to a flow-induced shear force. As the Brownian motion is significant for particles in the nm-size regime, the obtained trajectories can be understood as a superposition of a 2D stochastic motion (random walk) with a 1D deterministic motion (drift in direction of the flow).

We showed, for the first time, that such trajectories can be decomposed in both contributions, which provides information about the diffusion coefficient (of the random walk) and the drift velocity (of the deterministic motion). Interestingly, these information can, in turn, be used to accurately quantify the mobility of and the shear force acting on any of the tracked nanoparticles and thereby provide a new possibility to quantify nanoparticle properties. For example, as the determination of the shear force requires only knowledge of the trajectory but not of details of the system, we successfully used this approach to accurately determine the relationship connecting (hydrodynamic) nanoparticle size and the generated shear force. In this way, we were able to calibrate to flow conditions within the microfluidic channel based on experiments (instead of relying on theoretical assumptions), which is important for applications in flow-based force measurements (see below). Furthermore, this calibration also enabled us to determine the size distribution of nanoparticles with nm accuracy and single-particle resolution. If a component of the nanoparticles is stained with a fluorescent dye, this method, which we coined 2D flow nanometry, can be used to accurately quantify size-content relationships of nanoparticles (such as liposomes, exosomes, and viruses).
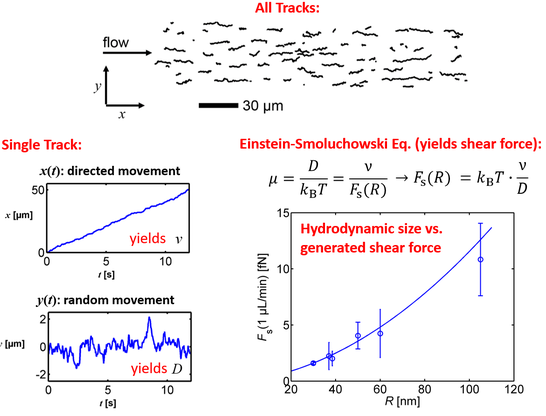
In addition to the acting shear force, the approach also provides accurate information on the mobility of the SLB-linked nanoparticle, which mainly depends on the particulars nanoparticle-SLB interaction, i.e., the linkers used for attaching the nanoparticles to the SLB. In fact, our previous work on multivalent model systems showed that the linking stoichiometry, i.e., the number of linkers that attach a nanoparticle of interest to the SLB, can be accurately determined from the mobility. Flow-based measurements offer a significant advantage in this context, as they simultaneously provide two measures for the mobility, the diffusion coefficient and the drift velocity, the latter of which can be determined with much higher accuracy.
Flow-based force measurements
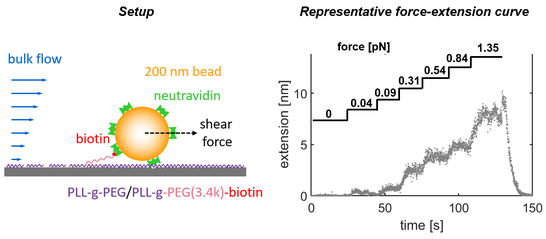
Nanoparticles that were directly linked to an immobile interface will not be able to move freely upon application of a shear force, but translate a certain fraction of this force into the linkers keeping the nanoparticle at the interface. As the generated shear force is precisely known from flow-based mobility studies (introduced above), this setup provides a new means to perform shear force-based single-molecule force measurements (similar to those done using atomic force microscopy (AFM) or optical/magnetic tweezers). If a macromolecule of interest is used to link the nanoparticle to the interface, this setup can be used to quantify the extension of this macromolecule in dependence of the applied shear force, yielding force-extension curves. We successfully applied this methodology to probe the nanomechanics of short polymer chains (as shown in the following figure), but also to study the unfolding and refolding of proteins.
Furthermore, if a ligand-receptor interaction of interest is part of the linking strategy (instead of using a covalent linkage), this setup can also be used to probe the dynamics of nanoparticle detachment in dependence of the applied shear force. Such measurements are conceptually similar to AFM-based force spectroscopy experiments and yield information about the energy landscape of the ligand-receptor interaction, but are done (in contrast to AFM) at negligible loading rates and therefore enable to probe small energy barriers that are inaccessible to complementary approaches.
In any case, since microscopy is used as readout, the fate of thousands of individual nanoparticles can be followed within a single experiment, thereby providing excellent measurement statistics even for short measurement times combined with the possibility to characterize heterogeneous samples. In our home-made setup we currently achieve sub-nm resolution in the measurement of force-extension curves of polymers or proteins, while the applicable force ranges typically between few fN up to 10 pN.
References
1. Block, S.*; Johansson Fast, B.; Lundgren, A.; Zhdanov, V.P.; Höök, F.* Two-dimensional flow nanometry of biological nanoparticles for accurate determination of their size and emission intensity. Nature Communications 7, 12956 (2016).
2. Lundgren, A.O.#; Johansson Fast, B.#; Block, S.; Agnarsson, B.; Reimhult, E.; Gunnarsson, A.; Höök, F.* Affinity purification of membrane proteins in native supported membranes. Nano Letters 18, 381-385(2018).
3. Li, J.H.; Santos-Otte, P.; Au, B.; Rentsch, J.; Block, S.; Ewers, H.*. Directed manipulation of membrane proteins by fluorescent magnetic nanoparticles. Nature Communications 11, 4259 (2020).
4. Kerkhoff Y, Azizi L, Mykuliak VV, Hytönen VP, Block S*. Microfluidics-based force spectroscopy enables high-throughput force experiments with sub-nm resolution and sub-pN sensitivity. Small 19, 202206713 (2023).
