Integrated transcriptomics reveal novel paradigms of splice site selection
RNA-Seq and downstream bioinformatics allow the transcriptome-wide quantification of splice site choices. By integrating multiple RNA-Seq datasets from siRNA-mediated perturbations of core components of the spliceosome, we have defined groups of spliceosomal proteins implicated in specific changes in alternative splice site choice. A multi-level bioinformatics analysis pipeline revealed common patterns or features within changed splicing events and led to our current focus on NAGNAG alternative splicing. We now use classical biochemical experiments to gain mechanistic insights into protein-specific changes in splice site selection. After careful validation of bioinformatics predictions using splicing-sensitive RT-PCRs we use bioinformatics-based mutations of cis-regulatory RNA sequences or structure-guided mutations of trans-acting factors for a detailed mechanistic analysis of alternative splice site selection. Ultimately, the combined bioinformatics and biochemical approaches will reveal new paradigms of splice site selection and contribute to understand the incredibly complex regulation of (alternative) splicing.
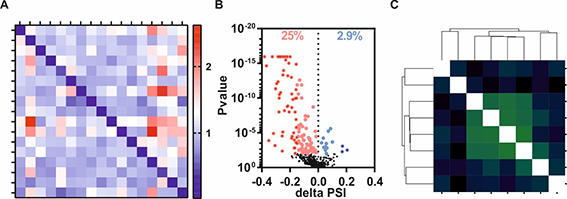
A, Heatmap confirming specific splicing factor knockdowns within RNA sequencing data.
B, Volcano blot revealing a directed effect of a splicing factor knockdown on alternative acceptor splice site choice (significant events are highlighted by colors).
C, Correlation matrix reveals co-regulation of a certain intron subtype by a subset of splicing factors.
