Viral protease inhibitors by fragment-based de novo design
Viral proteases cleave the viral polyprotein and thus represent a universal target for inhibiting viral replication. The major challenge is the selectivity of potential inhibitors over human proteases, which can be well addressed by computer-aided drug design. In a virtual screening campaign, structure-based design was used to find flaviviral NS2B-NS3 protease inhibitors (IC50 0.09 µM for the dengue NS2B-NS3 protease, ACS Med. Chem. Lett., 11:514-520, 2020).
Proteases offer the opportunity to pursue chemically novel approaches since selectivity can be achieved via systematic fragment-based design. In the first step, (covalently) binding fragments are identified, which are then expanded in the computer model based on their structure. Since the binding of the initial fragment can be validated by mass spectrometry, this approach is ideal for an experimentally driven, rational structure-based design of new ligands. For the enteroviral 3C protease (3CPro), we identified phenylthiomethylketone-based small fragments that bind covalently and selectively. The initially identified fragment was expanded with the aid of computers, which improved the activity as predicted (J. Med. Chem., 61:1218-1230, 2018).
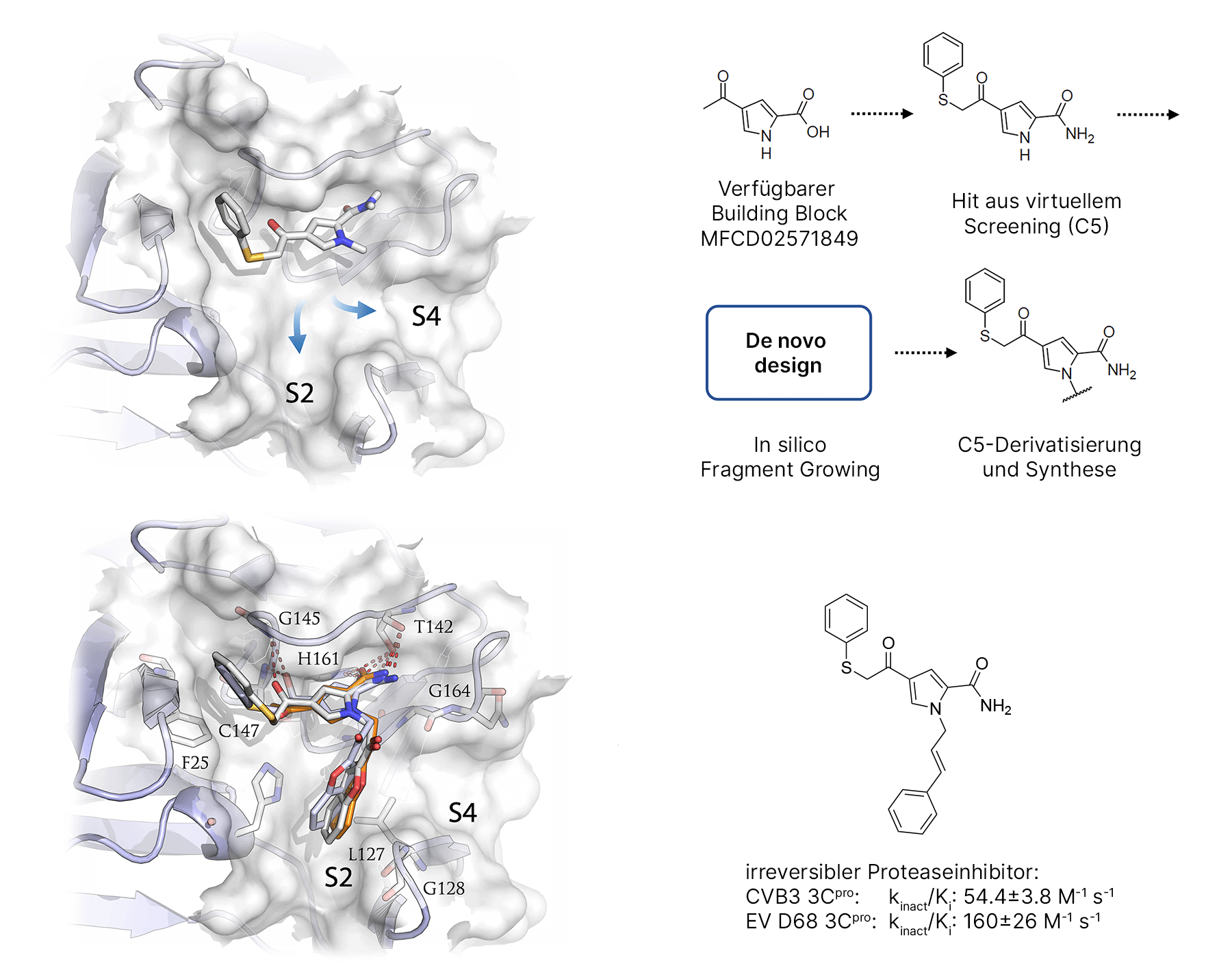
Fragment-based design strategy for irreversible ligands of the viral 3Cpro. Upper left: initial hit with exit vectors into the S3 and S4 binding pockets. Top right: starting fragment for the synthesis of the derivatives. Below: irreversible inhibitor
Links:
- p. Pach, T M Sarter, R Yousef, D Schaller, S Bergemann, C Arkona, J Rademann, C Nitsche, and G Wolber. Catching a moving target: comparative modeling of flaviviral NS2B-NS3 reveals small molecule Zika protease inhibitors, ACS Medicinal Chemistry Letters, 11(4):514-520, 2020 [doi:10.1021/acsmedchemlett.9b00629].
- R. Schulz, A Atef, D Becker, F Gottschalk, C Tauber, S Wagner, C Arkona, A Abdel-Hafez, H H Farag, J Rademann, and G Wolber. Phenylthiomethyl ketone-based fragments show selective and irreversible inhibition of enteroviral 3C proteases, J Med Chem, 61(3):1218-1230, 2018 [doi:10.1021/acs.jmedchem.7b01440].
- D. Becker, Z. Kaczmarska, C. Arkona, R. Schulz, C. Tauber, C. Wolber, G. Hilgenfeld, R. Hilgenfeld, M. Coll, and J. Rademann. Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding fragments, Nat Commun, 7:12761, 2016 [doi:10.1038/ncomms12761].