Tailored ligands for G-protein coupled receptors
GPCRs as signaling machinery
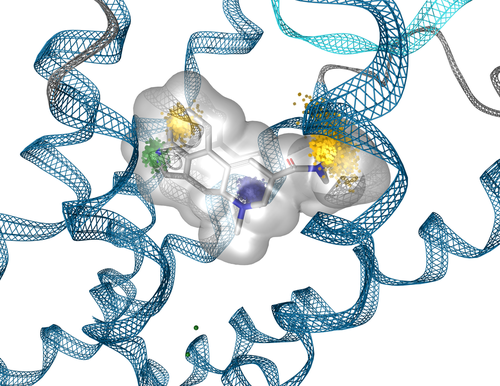
Dynophore von LSD im Serotonin-Rezeptor erklärt funktionelle Selektivität (biased Signaling)
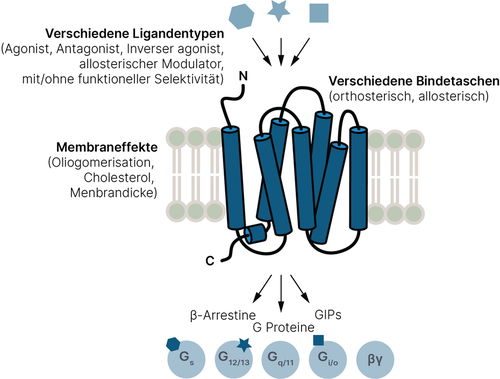
G protein-coupled receptors (GPCRs) are an integrative and highly dynamic signaling machinery that transmits chemically encoded information through multiple signaling pathways across the cell membrane (Bioorg. Med. Chem., 23:3907-3912, 2015, Trends Pharmacol. Sci., 35:630- 638, 2014). About 700 drugs (about 35% of all approved) act directly on 134 known GPCRs (Molecular Pharmacology, 93, 251-258, 2018) in a wide range of indications. Despite this high biological and pharmacological relevance, the underlying mechanisms related to receptor dynamics are poorly understood, which is due to the complex nature of GPCR signaling (Chem Rev, 117, 4-20, 2014, Fig. 1). Many partial GPCR agonists already on the market (e.g. pilocarpine for glaucoma therapy or buprenorphine as an analgesic) were not developed rationally, i. that is, the mechanism of the partial activation is not mechanistically understood.
The work of Brian Kobilka (Nobel Prize 2016) on the crystallization of the β‑adrenoceptors started a revolution in structural biology, through which 498 crystal structures of GPCRs are available today (as of January 2023) and which makes this receptor class accessible to structure-based, rational drug design. However, the receptor dynamics and the associated downstream signaling remain a major challenge, since crystal structures can never represent the conformational ensemble of the receptor. The biological effects caused by receptor dynamics relevant can only be assessed by molecular dynamics simulations. A mechanistic understanding of ligand-dependent receptor activation is essential for the rational design of GPCR modulators linked to a signaling profile essential for pharmacological action.
In recent years, our research group has specialized in developing models that consider GPCRs as dynamic units and take into account their complex signal repertoire. An important goal is to address the specific activation of a given pathway (functional selectivity/bias) in order to design tailor-made ligands for the activation of a given intracellular pathway. The strength of these models is a dynamic view of the receptor with special consideration of dynamic ligand binding patterns. For this purpose, we have developed a new modeling technique, dynamic 3D pharmacophores, which combines the advantages of classical structure-based design with the complexity of molecular dynamics simulation.
The long-term goal is the development of ligands with tailored functional selectivity. As model systems, we are initially focusing on muscarinic receptors and opioid receptors (Mol. Inf. 34:526-530, 2015, Bioorg. Med. Chem. Lett., 26:4769-4774, 2016, J. Med. Chem. 57:6739-6750, 2014, Molecules, 25:2087, 2020, Molecules, 26:3267, 2021, Pharmaceuticals, 15:680, 2022), since high therapeutic potential and sufficient data are available for these receptors. In cooperation with Prof. Dr. Holzgrabe (Würzburg) and Prof. Dr. Mohr/Prof. dr Weindl (Bonn) we were able to show on an atomistic-mechanical level how allosteric residues of bitopic ligands change the G protein recruitment profile and thus the intracellular signaling (ACS Chem. Biol., 12:1743-1748, 2017, ACS Pharmacol. Transl. Sci., 3:859-867, 2020). Independent of this effect, we were able to demonstrate the existence of ligand binding ensembles on the M2 receptor, in which a ligand can assume two different binding modes, one stabilizing the active and the other the inactive receptor state (J. Biol. Chem., 291:16375-16389, 2016).
Tailored functional selectivity at opioid receptors may reduce dose-limiting side effects such as respiratory failure (e.g., as reported for US-approved 2020 oliceridine (µOR)). A DFG-funded research project is currently running until 2024 for the development of safer analgesics using ligands with functional selectivity of the κ‑opioid receptor. Recently (Molecules, 28:718, 2023), we were able to develop a mechanistic model of the diterpenoid salvinorin A from Salvia divinorum at the κ‑opioid receptor, which will serve as the basis for novel, uncharged modulators.
We acknowledge and for financial support of this research area by: