Novel Computational Methods in Drug Design and Discovery
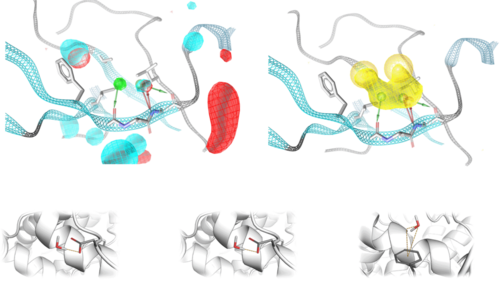
Pyrod uses solvation kinetics to sense interaction patterns for potential ligands
Image Credit: G. Wolber / D. Schaller
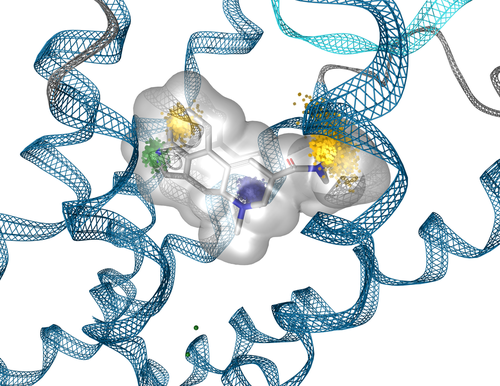
Dynophore von LSD im Serotonin-Rezeptor erklärt funktionelle Selektivität (biased Signaling)
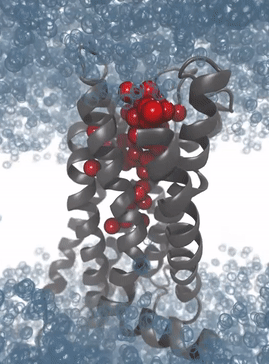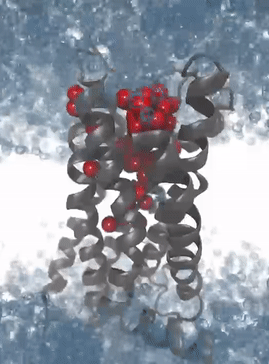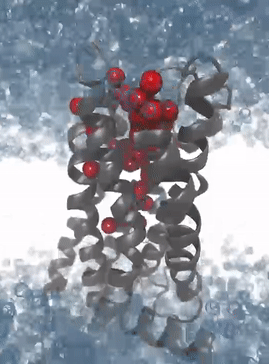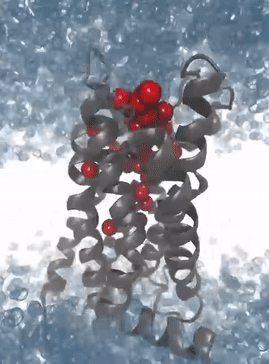
Simulation of the protein dynamics of the M2 receptor: The solvation patterns of the binding pocket enable the recognition of interaction patterns for the development of 3D pharmacophores for virtual high-throughput screening
With the increase in available computing power and the availability of General Purpose Computation on Graphics Processing Units (GPGPUs, calculations on graphics cards), molecular dynamics simulations have become routine in pharmaceutical drug design. The programs we recently developed Dynophores (dynamic 3D pharmacophore models) and PyRod allow the systematic analysis of large amounts of data from molecular dynamics simulations and their use for drug design and virtual screening. Especially for ligands of G protein-coupled receptors, the use of Dynophores is an important tool to understand and model small differences in ligand binding with the associated conformational change of the ternary complex and the associated intracellular signaling. BIth the analysis of increasing amounts of available data ('Big Data') and machine learning have become an integral part of design approaches in our working group.
In pharmaceutical-chemical research, the focus is primarily on the application of computer-aided methods for the development of new bioactive molecules, their characterization, synthesis, and optimization. In classical structure-based drug design, my working group has gained many years of experience in the application of virtual screening, 3D pharmacophores, docking, ensemble docking, and molecular dynamics simulations. New methods of artificial intelligence support our synthesis ideas, especially in de novo design. Through structure-informed learning, we combine the advantages of machine learning with the transparency of structure-based design, which is particularly important for understanding ligand-induced conformational changes for receptors.
Current Dynophore applications:
- M. Janežič, K. Valjavec, K. B. Loboda, B. Herlah, I. Ogris, M. Kozorog, M. Podobnik, S. G. Grdadolnik, G. Wolber, and A. Perdih. Dynophore-based approach in virtual screening: a case of human DNA Topoisomerase IIα, Int J Mol Sci, 22(24):13474, 2021. [doi:10.3390/ijms222413474]
- P. Durairaj, L. Fan, D. Machalz, G. Wolber, and M. Bureik. Functional characterization and mechanistic modeling of the human cytochrome P450 enzyme CYP4A22, Febs Lett, 593(16):2214-2225, 2019. [doi:10.1002/1873-3468.13489]
- J. Mortier, J. R. C. Prévost, D. Sydow, S. Teuchert, C. Omieczynski, M. Bermudez, R. Frédérick, and G. Wolber. Arginase structure and inhibition: Catalytic site plasticity reveals new modulation possibilities, Scientific Reports, 7(1):13616, 2017. Links: [doi:10.1038/s41598-017-13366-4]
- B. Nizami, D. Sydow, G. Wolber, and B. Honarparvar. Molecular insight on the binding of NNRTI to K103N mutated HIV-1 RT: molecular dynamics simulations and dynamic pharmacophore analysis, Mol Biosyst, 12:3385-3395, 2016. [doi:10.1039/C6MB00428H]
- A. Bock, M. Bermudez, F. Krebs, C. Matera, B. Chirinda, D. Sydow, C. Dallanoce, U. Holzgrabe, M. De Amici, M. J. Lohse, G. Wolber, and K. Mohr. Ligand Binding Ensembles Determine Graded Agonist Efficacies at a G Protein-Coupled Receptor, J Biol Chem, 291(31):16375-16389, 2016. [doi:10.1074/jbc.M116.735431]
Aktuelle PyRod-Anwendungen:
- S. Pach, T. M. Sarter, R. Yousef, D. Schaller, S. Bergemann, C. Arkona, J. Rademann, C. Nitsche, and G. Wolber. Catching a moving target: comparative modeling of flaviviral NS2B-NS3 reveals small molecule Zika protease inhibitors, ACS Medicinal Chemistry Letters, 11(4):514-520, 2020. [doi:10.1021/acsmedchemlett.9b00629]
- D. Schaller, and G. Wolber. Pyrod enables rational homology model-based virtual screening against MCHR1, Mol Inf, 39(6):e2000020, 2020. [doi:10.1002/minf.202000020]
Übersichtsartikel:
- Noonan, K. Denzinger, V. Talagayev, Y. Chen, K. Puls, C. A. Wolf, S. Liu, T. N. Nguyen, and G. Wolber. Mind the gap - deciphering GPCR pharmacology using 3D pharmacophores and artificial intelligence, Pharmaceuticals, 15(11):1304, 2022.[doi:10.3390/ph15111304]
- D. Schaller, D. Sribar, T. Noonan, L. H. Deng, T. N. Nguyen, S. Pach, D. Machalz, M. Bermudez, and G. Wolber. Next generation 3D pharmacophore modeling, WIREs Comp. Mol. Sci., 10(4):e1468, 2020. [doi: 10.1002/wcms.1468]