RNP remodeling enzymes
RNP remodeling enzymes
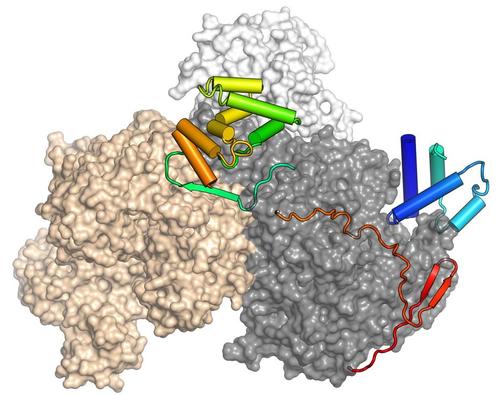
RNA-dependent nucleoside triphosphatases (NTPases) utilize the chemical energy of nucleoside triphosphate (NTP) hydrolysis to exert various biochemical activities. Due to the frequently observed ability to unwind RNA duplexes in vitro, these proteins are often referred to as "RNA helicases". However, in vivo they may also exert other functions, including RNA annealing, RNA clamping, buildup of RNPs, displacement of RNA-bound proteins or displacement of RNPs from RNAs. These enzymes are ubiquitous throughout the phylogenetic tree of life, as their activities are crucial for numerous cellular processes, including ribosome biogenesis, pre-mRNA processing, RNA transport or translation initiation. Based on sequence alignments, nucleic acid-dependent NTPases have been classified into six super-families (SFs). SF2 contains the largest number of members and has been subdivided into ten families, five of which contain RNA helicases (i.e. the DEAD-box, RIG-I-like, DEAH/RHA, NS3/NPH-II and Ski2-like families). DEAH/RHA and Ski2-like proteins are sometimes also grouped together as DExH-box enzymes. All SF2 enzymes share a common core composed of two RecA-like domains that couple NTP binding, hydrolysis and release of the products to nucleic acid or nucleic acid-protein complex binding or remodeling activities. These activities depend on a number of conserved sequence motifs that are more closely related within one family than between families. Enzymes grouped into the same family often share a number of additional domains that support and modulate the activities of the RecA domains. Yet further N-terminal and C-terminal regions or insertions are variable and specific for particular family members and can mediate interactions with substrates and cofactors or sub-cellular localization. We study DExH-box enzymes involved in pre-mRNA splicing and bacterial DExH-box enzymes. While the latter group of proteins has not been investigated extensively, they are thought to play important roles as co- or post-transcriptional gene regulators that facilitate the adaptation of bacteria to changing environments and stress conditions.
Recent publications
Pietrzyk-Brzezinska AJ, Absmeier E, Klauck E, Wen Y, Antelmann H, Wahl MC (2018) Crystal structure of the Escherichia coli DExH-box helicase HrpB. Structure 26, 1462-1473. (PubMed)
Absmeier E, Becke C, Wollenhaupt J, Santos KF, Wahl MC (2017) Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle 16, 100-112. (PubMed)
Henning LM, Santos KF, Sticht J, Jehle S, Lee CT, Wittwer M, Urlaub H, Stelzl U, Wahl MC, Freund C (2017) A new role for FBP21 as regulator of Brr2 helicase activity, Nucleic Acids Res, 13, 7922-7937. (PubMed)
Theuser M, Höbartner C, Wahl MC, Santos KF (2016) Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci USA 113, 7798-7803. (PubMed)
Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF, Wahl MC (2015) The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev 29, 2576-2587. (PubMed)