Bacterial transcription regulation
Bacterial transcription regulation
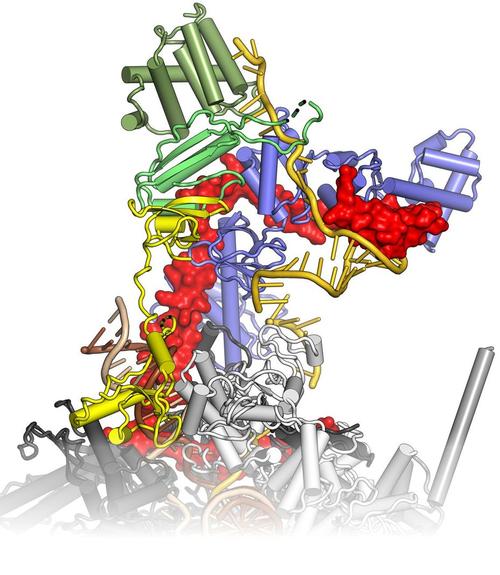
Bacteria transcribe their genomes with the help of multi-subunit RNA polymerases (RNAPs), which comprise two large β and β' subunits that form the active site, two regulatory α subunits and an ω subunit that supports RNAP assembly. The α2ββ’ω core enzyme cooperates with transcription factors and responds to signals on DNA templates and nascent RNAs to achieve full functionality in vivo. For example, elongating RNAP frequently enters an elemental paused state, and pausing can be stabilized by an RNA hairpin invading the RNA exit tunnel or by RNAP backtracking. RNA synthesis is terminated intrinsically, when the elongation complex transcribes a stable RNA hairpin followed by a uridine-rich stretch, or with the aid of transcription termination factor ρ. Pausing and termination can be further modulated by elongation factors, such as N-utilization substances A and G. Some regulatory factors or RNAs can stably insulate RNAP from the destabilizing effects of terminators over long distances (processive anti-termination). RNA-based processive anti-termination is exemplified by the polymerase utilization (put) signal of phage HK022. RNP-based processive anti-termination underlies the switching from immediate-early to delayed-early gene expression in other lambdoid phages as well as ribosomal RNA synthesis in Escherichia coli. In these processes, transcript-borne regulatory RNAs alone or in conjunction with protein factors assemble on the surface of RNAP and accompany the enzyme during further transcription by an RNA looping mechanism, rendering the elongation complex resistant to pause and/or termination signals downstream of the original modification site. We study the functional interplay of pausing, termination and continued transcription, which constitutes a pervasive gene regulatory principle in bacteria.
Recent publications
Huang YH, Said N, Loll B, Wahl MC (2019) Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis. Nucleic Acids Res, doi: 10.1093/nar/gkz290. (PubMed)
Krupp F, Said N, Huang YH, Loll B, Bürger J, Mielke T, Spahn CMT, Wahl MC (2019) Structural basis for the action of an all-purpose transcription anti-termination factor. Mol Cell 74, 143-157. (PubMed)
Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, Lee TC, Loll B, Behrmann E, Bürger J, Mielke T, Loerke J, Urlaub H, Spahn CMT, Weber G, Wahl MC (2017) Structural basis for λN-dependent processive transcription antitermination, Nat Microbiol 2, 17062. (PubMed)