Remus-Emsermann Group
Welcome to our lab
Plants leaves are colonised by complex microbial communities. Bacteria are the most dominant microorganisms on leaves. In our lab, we are mainly interested how bacterial species grow on plant leaves (the phyllosphere), which growth pattern they exhibit and how they interact with other species that share the leaf with them (figure 1). To do so, we use fluorescence microscopy techniques such as confocal laser scanning microscopy and widefield epifluorescence microscopy. By using bacteria that are genetically modified to produce fluorescent protein genes or bacteria are stained using fluorescence in situ hybridisation (FISH) and/or using fluorescent live/dead stains, we can determine the exact location of microbial cells on leaves and how they are interacting with each other. To analyse bacterial patterns we use image cytometry and spatial statistics that allow us to quantitatively assess aggregation and seggregation patterns of bacteria on leaves (figure 2).
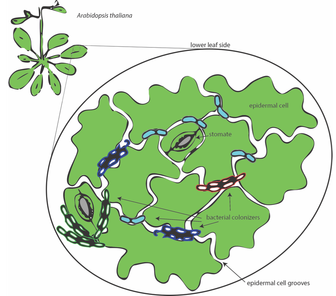
Figure 1 schematic of bacterial colonisation on the phyllosphere of Arabidopsis.
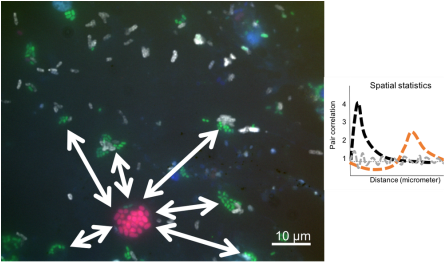
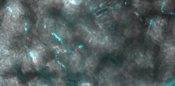
Figure 2 fluorescence microscopical image of (FISH-) fluorescently labelled bacteria that were recovered from environmentally grown Arabidopsis plants. Random dipoles were used to analyse spatial correlation resulting in pair correlation plots.
Since many leaf colonising bacteria cannot be used with the molecular toolbox available for E. coli and other model bacteria, a lot of research time in our lab is dedicated to adapt existing tools to make environmental bacteria more tractable to genetic manipulation (figure 3-7) and to effectively "make them shine". More information about "Chromatic bacteria toolbox" can be found here. This allows us to investigate bacteria in their natural habitat, such as the root nodules of soy beans (figure 4) or directly on plant leaves (figure 5).
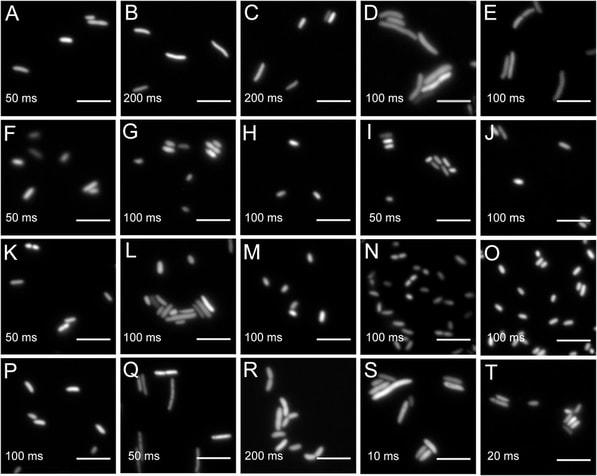
Widefield microscopy of environmental bacteria expressing fluorescent proteins. (A) E. coli DH5α (pMRE145). (B) E. coli DH5α::MRE-Tn5-145. (C) E. coli DH5α::MRE-Tn7-145. (D) Bradyrhizobium sp. Leaf396::MRE-Tn5-165. (E) Methylobacterium sp. Leaf92::MRE-Tn5-165. (F) Sphingomonas melonis FR1 (pMRE145). (G) S. melonis FR1::MRE-Tn5-145. (H) S. melonis FR1::MRE-Tn7-145. (I) Sphingomonas phyllosphaerae FA2 (pMRE135). (J) S. phyllosphaerae FA2::MRE-Tn5-145. (K) Erwinia amylovoraCFBP1430S (pMRE135). (L) E. amylovora CFBP1430S::MRE-Tn5-145. (M) E. amylovoraCFBP1430S::MRE-Tn7-145. (N) Pantoea agglomerans 299R (pMRE135). (O) P. agglomerans 299R::MRE-Tn5-145. (P) P. agglomerans 299R::MRE-Tn7-145. (Q) Pseudomonas citronellolis P3B5 (pMRE145). (R) P. citronellolis P3B5::MRE-Tn5-145. (S) Pseudomonas syringae pv. syringae B728a (pMRE145). (T) P. syringae B728a::MRE-Tn5-145. Exposure times used during image acquisition are depicted in the corresponding images. Scale bars represent 5 μm.
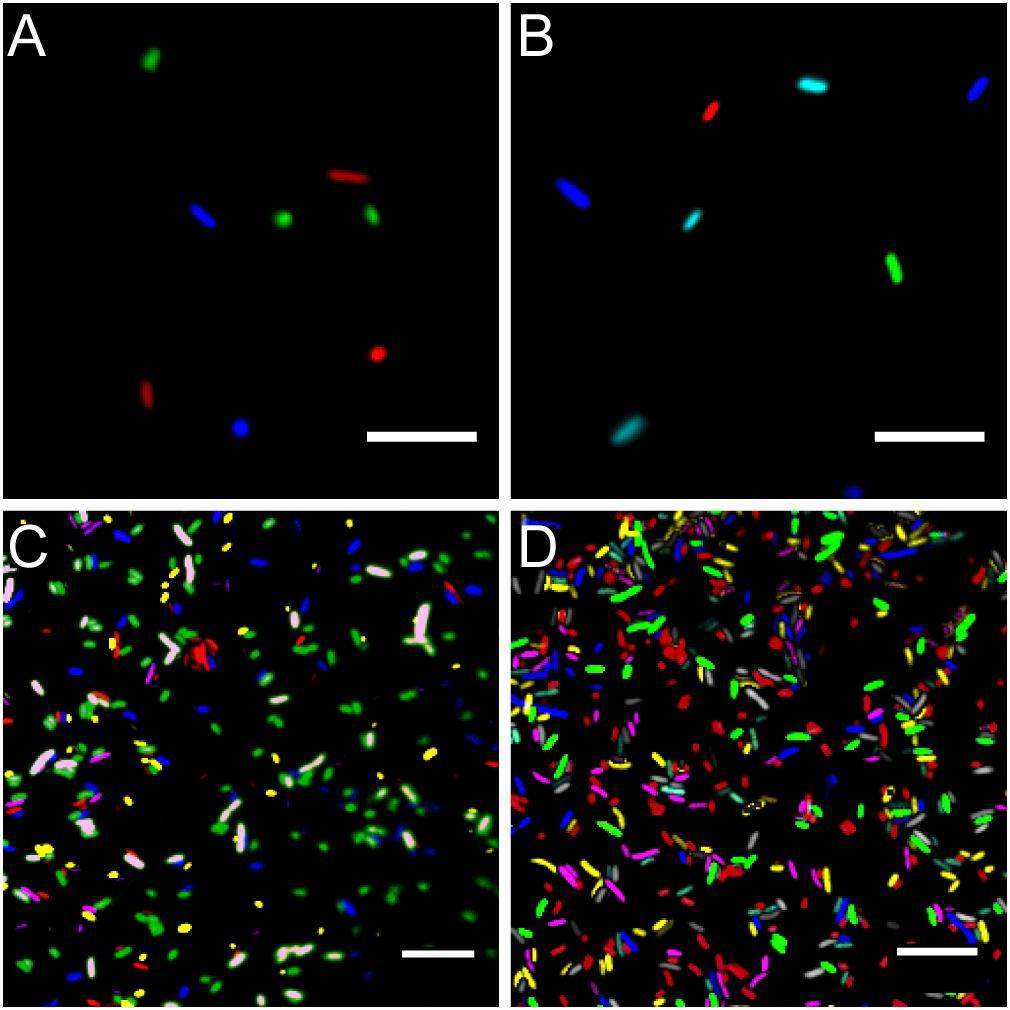 Microscopy images of fluorescent bacteria. (A) Widefield epifluorescence micrograph of E. coli expressing either mTB2 (blue), mCl3 (green), or mSc (red). (B) Widefield epifluorescence micrograph of E. coli expressing either mTB2 (blue), mTq2 (cyan), sYFP2 (green), or mSc (red). (C) Confocal microscopy of a mixed E. coli expressing either mTB2 (blue), mTq2 (magenta), mCl3 (yellow), mO2 (green), mSc (white), or mCa (red). (D) Confocal microscopy of a mixed E. coli expressing either mTB2 (cyan), mTq2 (magenta), sGFP2 (yellow), sYFP2 (gray), mO2 (red), mSc (green), or mCa (blue). In all cases, E. coli harboring pMRE14X plasmids series were used. Scale bar: 10 μm.
Microscopy images of fluorescent bacteria. (A) Widefield epifluorescence micrograph of E. coli expressing either mTB2 (blue), mCl3 (green), or mSc (red). (B) Widefield epifluorescence micrograph of E. coli expressing either mTB2 (blue), mTq2 (cyan), sYFP2 (green), or mSc (red). (C) Confocal microscopy of a mixed E. coli expressing either mTB2 (blue), mTq2 (magenta), mCl3 (yellow), mO2 (green), mSc (white), or mCa (red). (D) Confocal microscopy of a mixed E. coli expressing either mTB2 (cyan), mTq2 (magenta), sGFP2 (yellow), sYFP2 (gray), mO2 (red), mSc (green), or mCa (blue). In all cases, E. coli harboring pMRE14X plasmids series were used. Scale bar: 10 μm.

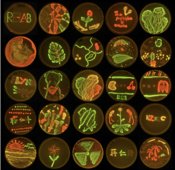
Figure 3 several phyllosphere isolates tagged with different constitutively expressed fluorescent proteins excited with UV light.
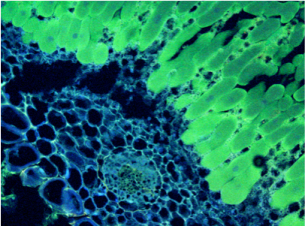
Figure 4 Soybean root nodule colonized by sYFP2 expressing Bradyrhizobium japonicum.
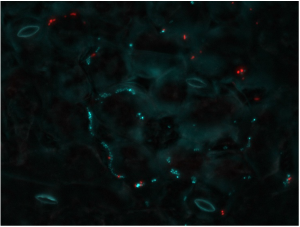
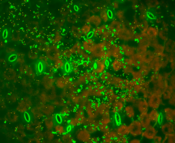
Figure 5 Arabidopsis leaf colonized by Pantoea agglomerans (in cyan) and Pseudomonas syringae DC3000 (in red)
Recently, our group has also started investigating the intricate interactions between herbivorous insects, leaf-associated bacteria, and their host plants. To study these tri-trophic relationship under controlled conditions, we have developed a gnotobiotic system that enables the cultivation of axenic (germ-free) caterpillars feeding on Arabidopsis thaliana inoculated with defined bacterial communities. This system allows us to trace the transmission and persistence of phyllosphere bacteria in the insect gut, assess their impact on insect development and performance, and understand how plant-associated microbiota influence plant–insect interactions.
To bring these insights closer to real-world conditions, we are expanding our experiments to include cabbage crops and their specialist Lepidoptera pests. Through this work, we aim to contribute to the development of microbiome-based strategies for sustainable crop protection.
Next to this main theme, we have been working on:
The interaction of phyllosphere bacteria with the herbivorous insects pest "great white cabbage butterfly" (Pieris brassicea).
Environmental studies to screen for antibiotic resistant bacteria,
the characterisation of oil degrading bacteria from plant leaves,
biocontrol of Erwinia amylovora on apple blossoms,
and the phyllosphere microbiota of kunzea in close vicinity of hydrothermal springs.