Research
Microbial proteomics, Thiol-redox proteomics and redox regulation in Gram-positive bacteria
Our research is based on proteomic, transcriptomic, biochemical and genetic approaches to study regulatory mechanisms, targeting, modifications, damage, aggregation and repair mechanisms of proteins in model Gram-positive bacteria, like Bacillus subtilis. Using gel-based proteomics we defined different subproteomic fractions such as extracellular, membrane- or cell wall-associated proteins to get insights into protein targeting and secretion mechanisms (1). The analysis of the protein synthesis profiles and the definition of proteomic signatures in response to defined stress or starvation conditions uncovered the mode of action of antimicrobial and toxic compounds (2). Furthermore, thiol-redox proteomic studies in response to oxidative and electrophile stress have been used to analyze the extent of reversible thiol modifications and protein damage in cytoplasmic proteins at a genome-wide scale (3, 7-9). These global analysis have identified novel redox-sensing regulators that specifically sense and respond to reactive electrophiles and strong oxidants, such as the MarR/DUF24-family repressor YodB, CatR and the positive regulator HypR (4, 5, 7). We further identified important targets for protein S-thiolations by strong oxidants like NaOCl in model Gram-positive bacteria, such as B. subtilis, Corynebacterium glutamicum, Mycobacterium smegmatis and in the major pathogen Staphylococcus aureus (7-10). Our current aims are to elucidate the physiological roles of NaOCl-sensitive thiol-switches, including S-bacillithiolations and S-mycothiolations under oxidative stress conditions in Gram-positive bacteria using genetic, biochemical and molecular biological methods.
Proteomics of protein secretion mechanism in bacteria
Using gel-based proteomics we defined different subproteomes of B. subtilis including the extracellular proteome, lipoproteome and cell wall proteome to analyze protein targeting and protein secretion mechanisms. In the course of several EU projects, we performed comparative extracellular and cell wall proteome analyses of B. subtilis mutants lacking components of the protein secretion machinery (e.g. targeting factors, Sec- and Tat-translocases, type-I and II signal peptidases, folding catalysts and proteases). Novel mechanisms of protein secretion were described for B. subtilis in collaboration with the Groningen secretion groups of Jan Maarten van Dijl and Sierd Bron. We identified substrates for the alternative Twin-arginine-translocation (Tat) pathway in pathogenic Gram-positive bacteria (Streptomyces scabies) und Gram-negative bacteria (Shigella flexneri, entoropathogenic E. coli and Pseudomonas aeruginosa) in collaboration with Tracy Palmer, Long-Fei Wu and Rome Voulhoux. In collaboration with Genencor International (Danisco) the secretion efficiency of B. subtilis production strains optimized for subtilisin production was analyzed.
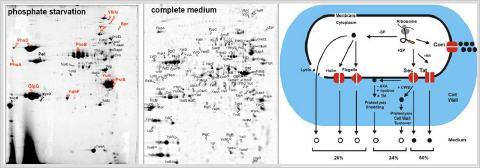
The extracellular proteome (A) and mechanisms for protein secretion in Bacillus subtilis (B). The extracellular proteome of B. subtilis consists of 113 extracellular proteins that are secreted under phosphate starvation or in LB medium. These include 54 predicted secretory proteins that possess N-terminal signal peptides (SP) and lack retention signals. The remaining 59 are unpredicted secreted proteins because these either lack an SP (17 cytoplasmic, 5 phage-related and 7 flagella-related proteins) or have retention signals for the cytoplasmic membrane (18 lipoproteins, 6 transmembrane proteins) or cell wall (6 cell wall proteins) (Antelmann et al., 2001; Tjalsma et al., 2004).
Global responses to Reactive Electrophilic Species (RES) in B. subtilis
Using systems biology approaches, such as transcriptomics, proteomics and metabolomics, global expression profiles were established in response to stress, starvation, antimicrobials and toxic electrophilic compounds, such as phenol, salicylic acid, chromanon, catechol, methylhydroquinone, diamide, formaldehyde, methylglyoxal and NaOCl for B. subtilis. We found that Reactive Electrophilic Species (RES) that are characterized by electron-deficient carbon centres (quinones, diamide and aldehydes) cause a signature that indicates depletions of thiol-redox buffers in the thiol metabolome. RES cause post-translational thiol-modifications in cytoplasmic proteins and the induction of specific protection mechanisms that are conserved in Gram-positive bacteria. Using thiol-redox proteomics and mass spectrometry it was shown that diamide and hypochlorite cause reversible thiol-modifications, including inter- and intramolecular protein disulfides and protein S-bacillithiolations in Bacillus species. In contrast, toxic quinones react with thiol-containing proteins predominantly via the thiol-(S)-alkylation chemistry leading to protein aggregation and depletion of the aggregated proteins from the proteome.
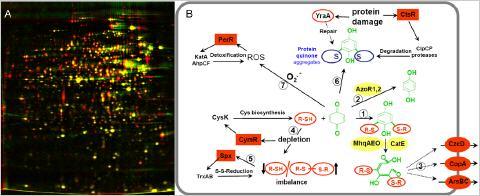
Depletion of thiol-containing proteins in the proteome by toxic quinones (A) and protective mechanisms against quinone toxicity in B. subtilis (B). A: The proteome of untreated B. subtilis cells (red image) is overlayed with the proteome after toxic quinone concentrations (green image). B: Quinones react with thiol redox buffers (R-SH) to quinone-S-adducts and cause an imbalanced thiol-redox homeostasis. The thiol-redox balance is restored by the derepression of the CymR regulon to increase cysteine biosynthesis and induction of the Spx-controlled thiol-disulfide reducing systems (TrxAB). Quinones are detoxified by the thiol-dependent dioxygenases MhqA, MhqE, MhqO and CatE or reduced by the azo-or quinone reductases AzoR1, AzoR2, YodC, MhqN. Quinones react also with protein thiols leading to protein-crosslinks or quinone-S-adducted proteins. These are proteolytically degraded by the CtsR-regulated ClpCP machinery. The oxidative mode of quinones leads to ROS production that upregulates the PerR-regulon.
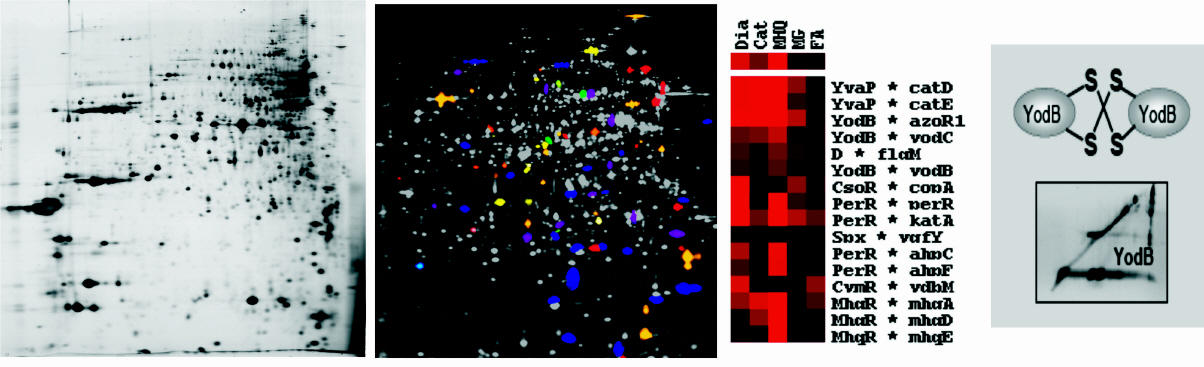
Characterisation of novel RES-sensing MarR/DUF24-family regulators in B. subtilis
B. subtilis is able to develop a specific adaptive response to quinone-like electrophiles and strong oxidants, such as diamide and NaOCl that is mediated by the MarR-type repressors MhqR, YodB, CatR, OhrR and HypR. The repressors MhqR, YodB and CatR control the expression of specific detoxification enzymes (azoreductases and and thiol-dependent dioxygenases) and OhrR controls the thiol-dependent peroxiredoxin OhrA. The MarR/DUF24 family repressors YodB and CatR sense diamide and quinones via intermolecular disulfide formation. In collaboration with Gottfried Palm and Winfried Hinrichs the crystal structures of the DUF24-family regulator HypR (YybR) was resolved that revealed structural changes upon oxidation. HypR controls positively the nitroreductase HypO (YfkO) in response to disulfide stress (diamide, NaOCl) and is activated by Cys14-Cys49' intersubunit disulfide formation.
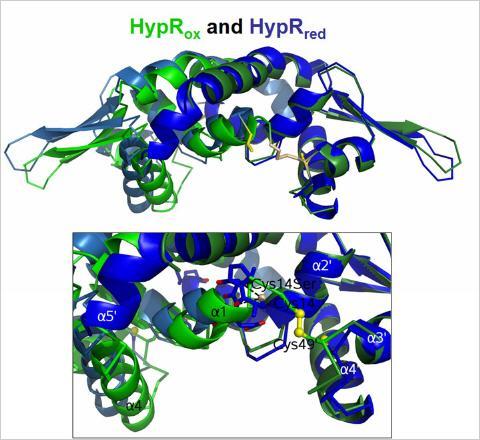
Structural changes of HypR upon oxidation. Superimposition of the oxidized HypR dimer (light and dark blue) and the reduced HypR dimer (light and dark green). Cys14 and Cys49' are shown as yellow sticks. The HypRox/red side view is shown and the structural changes in the Cys14-Cys49' pocket. One monomer of each dimer (on the right, dark blue and dark green) is aligned to visualize the differences in the opposing monomers.
The physiological role of the thiol redox buffers bacillithiol and mycothiol
Hypochlorite is a strong oxidant that is released by activated macrophages during infections with the aim to kill pathogenic bacteria. We have studied changes in the transcriptome and thiol-redox proteome caused by the strong oxidant hypochloric acid (NaOCl) in the model bacteria B. subtilis and Corynebacterium glutamicum. We discovered important roles of the bacillithiol (Cys-GlcN-Malate, BSH) redox buffer in B. subtilis in redox regulation and protection of active site Cys residues of conserved enzymes against irreversible oxidations under NaOCl stress conditions. NaOCl stress caused wide-spread S-bacillithiolation of the redox-sensing MarR-type repressor OhrR and of several amino acid biosynthetic enzymes, translation factors and antioxidant proteins in Bacillus species. In eukaryotes, protein S-glutathionylation has emerged as a major cellular regulatory mechanism and the inactivation of several metabolic enzymes is caused by S-glutathionylation in response to oxidative stress. Thus, our current research is directed to investigate the physiological role of BSH in redox regulation of cytoplasmic proteins by S-bacillithiolation among Firmicutes bacteria. Recently, we further identified 26 proteins in C. glutamicum that form mixed mycothiol protein disufides (S-mycothiolations) under hypochlorite stress conditions. These proteins include metabolic enzymes, translation factors and ribosomal proteins and peroxiredoxins. Thus, the role of S-mycothiolation in redox control and protein protection under infection-related conditions in pathogenic Corynebacteria is our future research focus.
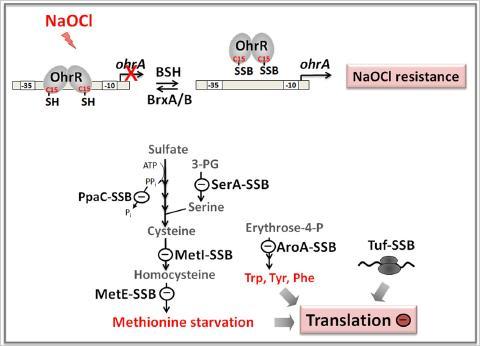
Protein S-bacillithiolation functions to protect proteins against overoxidation. We identified several conserved protein S-bacillithiolations across Firmicutes bacteria specifically after NaOCl stress. In B. subtilis, NaOCl leads to S-bacillithiolation of the OhrR repressor and several enzymes for Cys and Met biosynthesis. S-bacillithiolation of OhrR causes induction of the OhrA peroxiredoxin that confers NaOCl resistance. S-bacillithiolation of MetE and other amino acid biosynthetic enzymes as well as translation factors leads to methionine auxotrophy to stop translation during the time of NaOCl detoxification.
