Multivalent Interactions
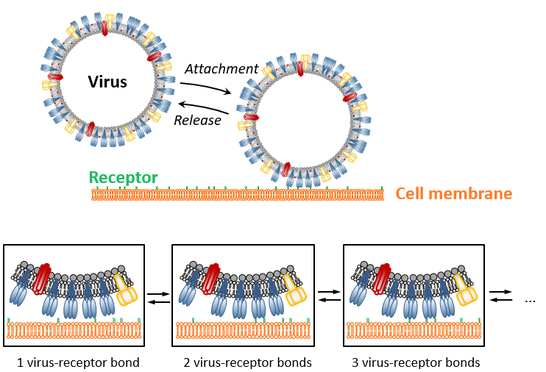
Multivalent interactions, i.e., multiple, non-covalent bonds acting in parallel, are typical for a multitude of biological processes, such as the attachment of viruses to the membrane of host cells. To elucidate multivalency-related processes in detail, it is inevitable to quantify the valency (i.e., the number of single interactions engaged in parallel) on the level of a single interaction complex (i.e., with single virus resolution). This is still a major challenge as the individual interaction is typically weak and therefore maintained only transiently, causing temporal fluctuations of the valency value and generating relatively broad valency distributions across the probed ensemble.
Quantification of multivalent interactions therefore requires methods that on the one hand allows for the characterization of many interaction complexes in parallel (while keeping, for example, the single-virus resolution) but also provide, on the other hand, sufficient temporal resolution to follow temporal fluctuations of the valency. To this end, we apply advanced microsopic techniques (e.g., total internal reflection fluorescence (TIRF) microscopy, confocal and scattering microscopy, atomic force microscopy-based force spectroscopy) to quantify the action of multivalent interactions in complementary settings. For example, equilibrium binding properties (equilibrium reaction rates) can be studied using binding assays based on TIRF and scattering microscopy, while non-equilibrium features (meachanical stability) are provided by atomic force microscopy.
Probing Virus Binding Using Total Internal Reflection Fluorescence (TIRF) microsopy
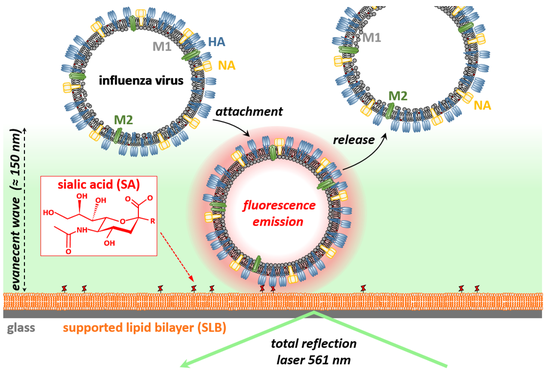
In this assay, the transient attachment and release of viruses is measured using TIRF microscopy. To this end, a supported lipid bilayer (SLB), which is supplemented with attachment factors of the virus of interest, is formed at an interfaces. The transient interaction of viruses with this artificial cell membrane is monitored using total internal reflection fluorescence (TRIF) microscopy, which excites only fluorescently labeled viruses being within 100 - 150 nm to the interface. In this way, viruses are only visualized by the microscope when bound to the artificial cell membrane. The following video shows a representative example, in which the transient interaction of influenza A viruses (strain X31; bright dots in the video) with a sialic acid-presenting, POPC-based SLB was probed.

In such experiments, the artificial cell membrane is in a fluid phase and hence, the viruses are able to move laterally even when bound to the membrane. Interestingly, the mobility of SLB-linked particles (diameter < 400 nm) mainly depends of the number of engaged lipids and enables to estimate the valency of a particular virus by quantifying its mobility (e.g., its diffusion coefficient). Application of single particle tracking to such movies therefore enables to obtain 3 key properties of multivalent interactions: i) the rate of virus attachment to the membrane, ii) a measure of the valency distribution of the viruses, and iii) the valency-dependent off-rate distribution.While the off-rate distribution can be quantified in absolute units, the rate of virus attachment and the valency value can currently only be estimated in qualitative terms. This shortcoming reflects lack of knowledge regarding key properties of the system (i.e., quantitative knowledge of the mobility of nm-sized membrane inclusions in SLBs and of the virus composition) and limitations in the instrumentation (in particular in the temporal resolution of the imaging process). Nevertheless, we successfully applied this assay to determine these binding properties for various viruses, including influenza A virus strains and the polyoma viruses SV40, and to assess how antivirals modify virus-receptor interactions (by probing changes in these key properties induced by addition of the drug).
References
1. Block S.*, Zhdanov VP, Höök F.* Quantification of multivalent interactions by tracking single biological nanoparticle mobility on a lipid membrane. Nano Letters16, 4382-4390 (2016).
2. Block S.* Brownian motion at lipid membranes: A comparison of hydrodynamic models describing and experiments quantifying diffusion within lipid bilayers. Biomolecules 8, 30 (2018).
3. Müller M, Lauster D, Wildenauer HHK, Herrmann A, Block S*. Mobility-based quantification of multivalent virus-receptor interactions: New insights into influenza A virus binding mode. Nano Letters 19, 1875-1882 (2019).
4. Wallert M, Nie C, Anilkumar P, Abbina S, Bhatia S, Ludwig K, Kizhakkedathu JN, Haag R*, Block S*. Mucin-inspired, high molecular weight virus binding inhibitors show biphasic binding behavior to influenza A viruses. Small 16, 2004635 (2020).
5. Nie C, Stadtmüller M, Prashad B, Wallert M, Ahmadi V, Kerkhoff Y, Bhatia S, Block S*, Cheng C*, Wolff T*, Haag R*. Heteromultivalent topology-matched nanostructures as potent and broad-spectrum influenza A virus inhibitors. Science Advances 7, eabd3803 (2021).
6. Hamming PHE, Overeem NJ, Diestelhorst K, Fiers T, Tieke M, Vos G, Boons GJPH, van der Vries E, Block S*, Huskens J*. Receptor density-dependent motility of influenza virus particles on surface gradients. ACS Appl. Mater. Interfaces 15, 25066-25076 (2023).
Probing Binding Strengths using Atomic Force Microscopy
One of the most important techniques to probe biomolecular interactions is atomic force microscopy (AFM), which directly measures the force needed to rupture an individual interaction. In this setting, one interaction partner is immobilized at a surface, while the other one is linked to a nm-sharp tip of a cantilever, which can be used to bring both interaction partners into contact (for a defined period of time) and to record the force needed to separate the interaction partners. This procedure generates force-separation curves with high force (~ 10 pN) and spatial (~ 1 nm) resolution. Typically, the force is continuously increased at a certain loading rate until rupture of the interaction is detected. Theoretical assessment of this scheme shows that the observed rupture force depends of the loading rate applied to the interaction and that the distribution of rupture forces, measured at different loading rates, contains information about the energetics of the interaction.
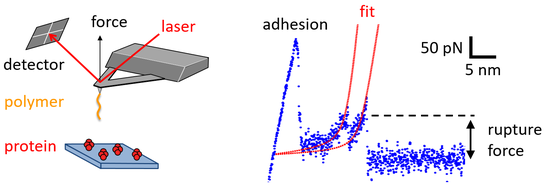
In recent works, we applied this methodology to quantify the interactions arising between proteins and biopolymers, such as polysaccharides and glycosaminoglycans, and investigated the impact of polymeric charge density on the protein-polymer interaction. Currently, we apply various AFM-based measurement schemes, including quantitative imaging in liquid environments and force spectroscopy, to study the properties of viral binding proteins. For non-enveloped viruses, the viral binding proteins are typically water-soluble and can be directly immobilized at either AFM tip of surface. In contrast, most viral binding proteins of enveloped viruses are membrane proteins and hence poorly soluble in water. In order to characterize the full-length protein (and not only its receptor-binding domain) and to mimic the lipid environment of the viral envelope, we develop protocols that enable to reconstitute binding protein of enveloped viruses into liposomes (see also the Reseach Topic Biomimetic Systems). These proteoliposomes can be used to generate lipid bilayers that present the viral protein of interest to receptor-presenting AFM tips and hence to conduct AFM-based force spectroscopy of virus-receptor interactions.
References
1. Block S*, Greinacher A, Helm CA, Delcea M*. Characterizing bonds formed between platelet factor 4 and negatively charged drugs using single molecule force spectroscopy. Soft Matter 10, 2775-2784 (2014).
2. Bally M, Block S, Höök F*, Larson G*, Parveen N, Rydell R. Physicochemical tools for studying virus interactions with targeted cell membranes in a molecular and spatiotemporally resolved context. Analytical Bioanalytical Chemistry 413, 7157-7178 (2021).


