Biomimetic model systems
Investigations involving biological specimen are often complicated by the fact that most systems of interest exhibit a significant compositional complexity. Often, the impact of the composition on the behavior of the biological system is not known and in fact subject to current research. An essential aspect of our work is therefore the preparation and characterization of model systems of defined compositional complexity, which are then used in our methods in order to accurately quantify functional aspects of macromolecules, such as the activity of single enzymes and channels or the interaction dynamics of (viral) proteins that bind to or release from biomimetic models of cell membranes. Typically, we generate such model systems by incorporation of macromolecules of interest into liposomes, yielding proteoliposomes (generated by protein reconstitution procedures) or native-membrane containing liposomes (generated by incorporation of membrane preparations into liposomes).
Activity of single enzymes, channels and porins
These investigations are conducted using proteoliposomes, which are generated by reconstitution of proteins of interest into liposomes. In order to probe the activity of the proteins, the proteoliposomes are loaded with a reporter dye, the fluorescence properties of which depend on the concentration of a relevant ion species. For example, for proteins that actively or passively translocate protons (pumps and channels, respectively), we encapsulate pH-sensitive dyes such as pyranine for proteins, which allow to quantify the luminal pH value of the proteoliposomes and hence the dynamics of protein activity by following changes in the luminal pH value over time.
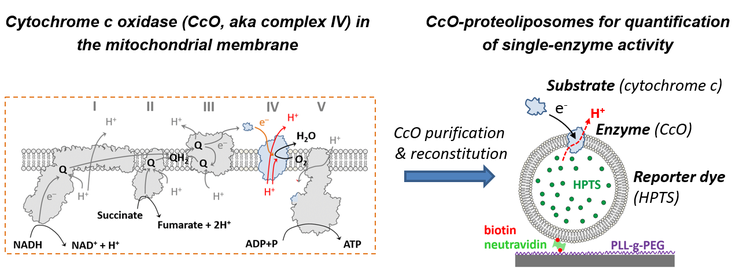
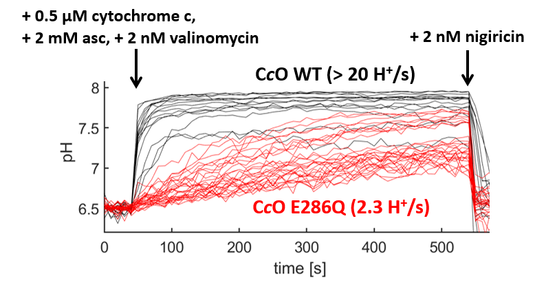
As proteoliposomes show a notable variation in size and typically vary with respect to the number of reconstituted proteins, accurate quantification is only possible by probing the response on the level of single proteoliposomes. To this end, we link the proteoliposomes to an interface and take advantage of the high resolution and sensitivity of current microscopes in order to determine the luminal pH value of individual proteoliposomes. An example of this approach is shown in the following video, in which only a small fraction of the liposomes have been equipped with a proton pump (the cytochrome c oxidase from Rhodobacter spheroids). Hence, only a fraction of the liposomes show a time-dependent change in fluorescence intensity (reflecting a change in luminal pH value of these proteoliposomes), while most of the liposomes do not contain the proton pump and hence do not show any changes in fluorescence intensity.

As thousands of liposomes can be probed in parallel, hundreds of proton uptake traces can be extracted from a single experimental run, which enables to quantify proton turnover rates with high statistical power and to resolve heterogeneities in the function across the ensemble.
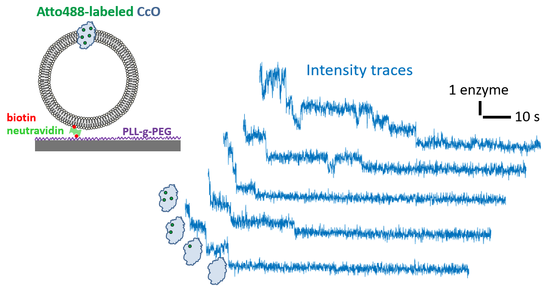
In the past, we successfully applied this methodology to quantify proton translocation by proton pumps and pH-activated proton channels as well as the permeability of porins for certain compounds. For these measurements, we used liposomes of various compositions, ranging between well-defined mixtures of synthetic or purified lipids to lipid mixtures that have been extracted from native sources (such as bacteria or plants). In order to determine the distribution of the proteins across the ensemble we reconstitute dye-labeled protein into liposomes, which do not carry a reporter dye. Owing to the single-molecule resolution of our microscopes, such samples can be subjected to stepwise bleaching experiments (see the following figure) that enable to determine the number distribution of proteins across the liposomes.
References
1. Berg, J.#; Block, S.#; Höök, F.; Brzezinski, P. Single proteoliposomes with E. coli quinol oxidase: proton pumping without transmembrane leaks. Israel Journal of Chemistry 57, 437-445 (2017).
2. Hugentobler, K.G.; Heinrich, D.; Berg, J.; Heberle, J.; Brzezinski, P.; Schlesinger, R.; Block, S.*. Lipid Composition Affects the Efficiency in the Functional Reconstitution of the Cytochrome c Oxidase. International Journal of Molecular Sciences 21, 6981 (2020).
Biomimetic model systems for binding studies
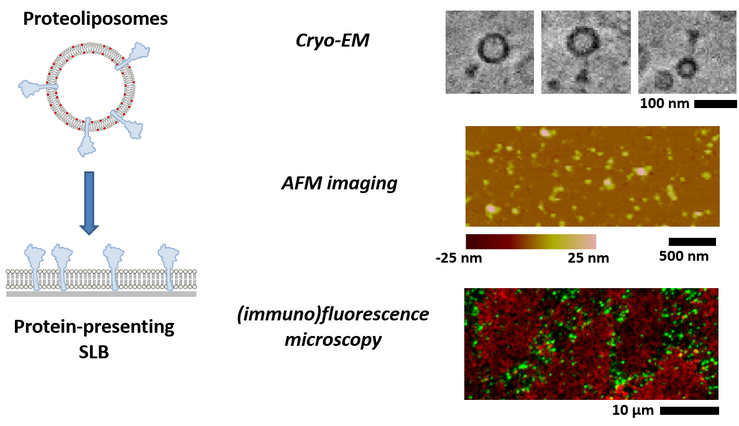
A major challenge in the quantification of virus-membrane interactions is the fact that viruses as well as cell membranes typically show a significant compositional complexity. In order to probe the impact of compositional complexity on the dynamics of virus binding, we develop biomimetic model systems of cell membranes, which are formed either using (i) proteoliposome or (ii) native-membrane containing liposomes. In the first approach, protein-based attachment factors (such as the transmembrane protein ACE2 in studies of coronavirus binding) are reconstituted into liposomes of known composition, providing full control of the final bilayer composition. In the second approach, vesicles are generated by mechanical disruption of cells. SLBs can typically not be formed using these vesicles alone but after supplementing them with synthetic liposomes, which tend to promote SLB formation. In this way, it is possible to generate SLBs that are enriched with native material (originating from cell membranes), but at the expense that the composition is much less well-defined as in the case of proteoliposomes. Hence, in parallel to developing such preparation approaches, our current work also includes the application of biochemical and biophysical approaches such as cryo-electron microscopy, atomic force microscopy, and immunofluorescence microscopy in order to characterize the generated SLBs with respect to surface concentration and distribution of relevant macromolecules.
References
1. Lundgren, A.O.#; Johansson Fast, B.#; Block, S.; Agnarsson, B.; Reimhult, E.; Gunnarsson, A.; Höök, F.* Affinity purification of membrane proteins in native supported membranes. Nano Letters 18, 381-385(2018).
2. Nie, C.; Stadtmüller, M.; Prashad, B.; Wallert, M.; Ahmadi, V.; Kerkhoff, Y.; Bhatia, S.; Block, S.*; Cheng, C.*; Wolff, T.*; Haag, R.* Heteromultivalent topology-matched nanostructures as potent and broad-spectrum influenza A virus inhibitors. Science Advances 7, eabd3803 (2021).


