Research
Numerous fundamental cellular processes, including virtually every aspect of gene expression and regulation, depend on the cooperation of RNAs and proteins. Specific RNAs and proteins associate to build up some of the most complex macromolecular machineries of living cells, such as ribosomes and spliceosomes, which mediate particular steps of gene expression. RNAs and proteins can also engage in more transient interactions, for example during co-transcriptional regulation of RNA polymerases or during post-transcriptional regulation of (pre-)mRNA life cycles. Moreover, RNA-protein complexes (RNPs) provide glimpses at the molecular ancestry of modern cells, which most likely evolved from an RNA-dominated world.
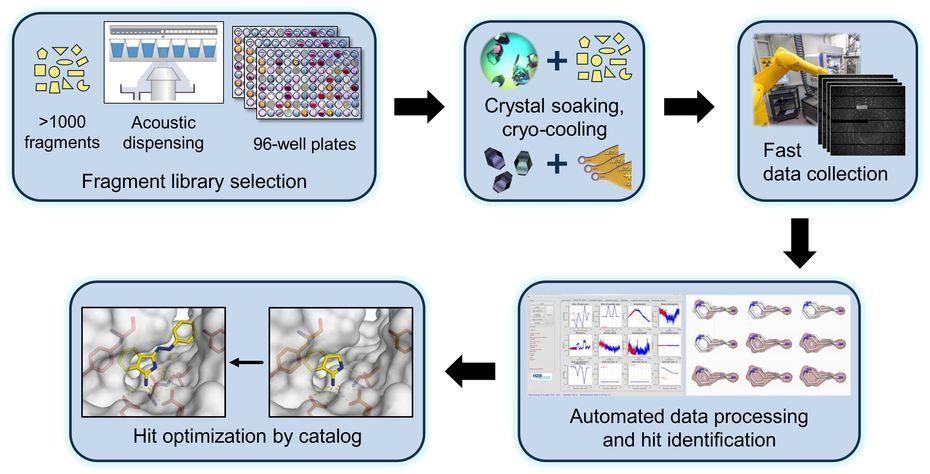
We investigate the molecular mechanisms, by which RNAs and proteins cooperate to bring about the biological functions of selected RNPs involved in transcription and pre-mRNA splicing. We also study selected RNP-remodeling enzymes as drivers of molecular RNP machineries and as mediators of co- and post-transcriptional gene regulation. Our group uses structural biochemical techniques, primarily macromolecular crystallography (MX) and single-particle cryo-electron microscopy (cryoEM), to elucidate 3D structures of RNPs or their components. Based on the structures, we conduct targeted functional analyses in vitro and in vivo. We also closely collaborate with the Joint Research Group MX of Helmholtz-Zentrum Berlin and Freie Universität at the local BESSY II storage ring, where we aim at exploiting MX-based fragment screening to develop small molecule probes and modulators of RNP function. We are a member of the Joint Berlin MX-Laboratory.
Bacterial transcription regulation
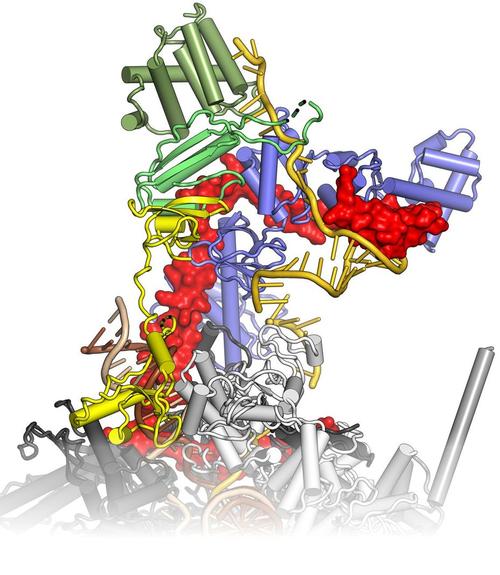
Bacteria transcribe their genomes with the help of multi-subunit RNA polymerases (RNAPs), which comprise two large β and β' subunits that form the active site, two regulatory α subunits and an ω subunit that supports RNAP assembly. The α2ββ’ω core enzyme cooperates with transcription factors and responds to signals on DNA templates and nascent RNAs to achieve full functionality in vivo. For example, elongating RNAP frequently enters an elemental paused state, and pausing can be stabilized by an RNA hairpin invading the RNA exit tunnel or by RNAP backtracking. RNA synthesis is terminated intrinsically, when the elongation complex transcribes a stable RNA hairpin followed by a uridine-rich stretch, or with the aid of transcription termination factor ρ. Pausing and termination can be further modulated by elongation factors, such as N-utilization substances A and G. Some regulatory factors or RNAs can stably insulate RNAP from the destabilizing effects of terminators over long distances (processive anti-termination). RNA-based processive anti-termination is exemplified by the polymerase utilization (put) signal of phage HK022. RNP-based processive anti-termination underlies the switching from immediate-early to delayed-early gene expression in other lambdoid phages as well as ribosomal RNA synthesis in Escherichia coli. In these processes, transcript-borne regulatory RNAs alone or in conjunction with protein factors assemble on the surface of RNAP and accompany the enzyme during further transcription by an RNA looping mechanism, rendering the elongation complex resistant to pause and/or termination signals downstream of the original modification site. We study the functional interplay of pausing, termination and continued transcription, which constitutes a pervasive gene regulatory principle in bacteria.
Recent publications
Huang YH, Said N, Loll B, Wahl MC (2019) Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis. Nucleic Acids Res, doi: 10.1093/nar/gkz290. (PubMed)
Krupp F, Said N, Huang YH, Loll B, Bürger J, Mielke T, Spahn CMT, Wahl MC (2019) Structural basis for the action of an all-purpose transcription anti-termination factor. Mol Cell 74, 143-157. (PubMed)
Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, Lee TC, Loll B, Behrmann E, Bürger J, Mielke T, Loerke J, Urlaub H, Spahn CMT, Weber G, Wahl MC (2017) Structural basis for λN-dependent processive transcription antitermination, Nat Microbiol 2, 17062. (PubMed)
Pre-mRNA splicing
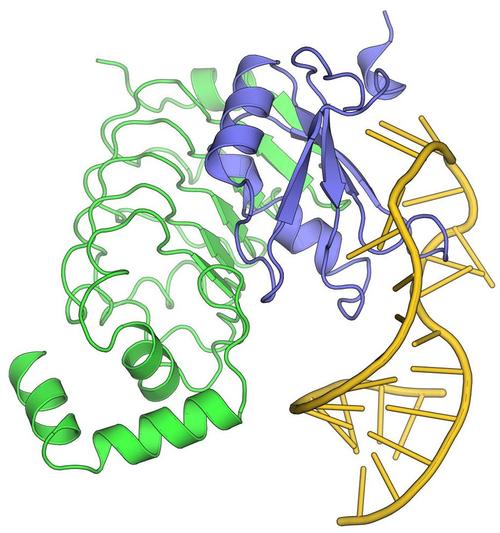
Many eukaryotic precursor messenger RNAs (pre-mRNAs) bear coding regions (exons) interspersed with non-coding intervening sequences (introns). Production of mature mRNAs thus requires pre-mRNA splicing, during which introns are excised and neighboring exons are ligated. A splicing reaction consists of two SN2-type transesterification reactions. In the first half-reaction, the phosphodiester bond at the 5’-splice site is attacked by the 2’-hydroxyl of a conserved adenosine of the branch point sequence in the intron, generating a free 5’-exon and an intron lariat-3’-exon. In the second half-reaction, the 3’-hydroxyl of the 5’-exon attacks the phosphodiester bond at the 3’-splice site, leading to exon ligation and excision of the lariat intron. Splicing is carried out by spliceosomes, which are large and highly dynamic molecular RNP machines. Spliceosomes are composed by five small nuclear (sn) RNPs (U1, U2, U4, U5 and U6 in the case of the major/U2-type spliceosome) and many additional non-snRNP splicing factors. For each round of splicing, a spliceosome is assembled, catalytically activated and, after splicing catalysis, disassembled. Each assembly, activation, catalysis and disassembly step is associated with profound compositional and conformational remodeling of the underlying RNA-protein interaction network. Higher eukaryotic genes typically contain more than one intron and their pre-mRNAs can undergo alternative splicing, giving rise to mature transcripts with different combinations of exons or portions of exons and thus ultimately to more than one protein. Alternative splicing is essential for producing the large repertoires of proteins required in higher organisms from a limited number of protein-coding genes, endows eukaryotes with entirely novel mechanisms to regulate their gene expression and supports novel principles of evolution. Splicing and alternative splicing are also of tremendous medical interest, as mutations in splicing factor genes have been associated with more than 300 disorders in humans and as at least 25 % of all inherited human diseases root in aberrant splicing. We are interested in how the molecular design principles of spliceosomes ensure the reliable identification of authentic splice sites, yet at the same time provide sufficient functional flexibility for alternative splicing.
Recent publications
De Bortoli F, Neumann A, Kotte A, Timmermann B, Schüler T, Wahl MC, Loll B, Heyd F (2019) Increased versatility despite reduced molecular complexity – evolution, structure and function of metazoan splicing factor PRPF39. Nucleic Acids Res, doi: 10.1093/nar/gkz243. (PubMed)
Weber G, DeKoster GT, Holton N, Hall K, Wahl MC (2018) Molecular principles underlying dual RNA specificity in the Drosophila SNF protein. Nat Commun 9, 2220. (PubMed)
Ulrich A, Seeger M, Schütze T, Bartlick N, Wahl MC (2016) Scaffolding in the spliceosome via single α-helices. Structure 24, 1972-1983. (PubMed)
Ulrich AKC, Schulz JF, Kamprad A, Schütze T, Wahl MC (2016) Structural basis for the functional coupling of the alternative splicing factors Smu1 and RED. Structure 24, 762-773. (PubMed)
Liu S, Mozaffari-Jovin S, Wollenhaupt J, Santos KF, Theuser M, Dunin-Horkawicz S, Fabrizio P, Bujnicki JM, Lührmann R, Wahl MC (2015) A composite, double-stranded/single-stranded RNA-binding region in the Prp3 protein important for U4/U6•U5 tri-snRNP stability and splicing. eLife 4, e07320. (PubMed)
RNP remodeling enzymes
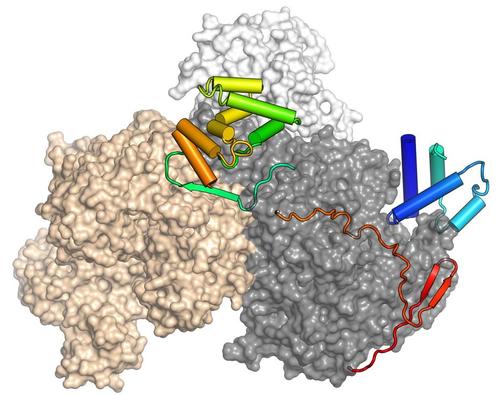
RNA-dependent nucleoside triphosphatases (NTPases) utilize the chemical energy of nucleoside triphosphate (NTP) hydrolysis to exert various biochemical activities. Due to the frequently observed ability to unwind RNA duplexes in vitro, these proteins are often referred to as "RNA helicases". However, in vivo they may also exert other functions, including RNA annealing, RNA clamping, buildup of RNPs, displacement of RNA-bound proteins or displacement of RNPs from RNAs. These enzymes are ubiquitous throughout the phylogenetic tree of life, as their activities are crucial for numerous cellular processes, including ribosome biogenesis, pre-mRNA processing, RNA transport or translation initiation. Based on sequence alignments, nucleic acid-dependent NTPases have been classified into six super-families (SFs). SF2 contains the largest number of members and has been subdivided into ten families, five of which contain RNA helicases (i.e. the DEAD-box, RIG-I-like, DEAH/RHA, NS3/NPH-II and Ski2-like families). DEAH/RHA and Ski2-like proteins are sometimes also grouped together as DExH-box enzymes. All SF2 enzymes share a common core composed of two RecA-like domains that couple NTP binding, hydrolysis and release of the products to nucleic acid or nucleic acid-protein complex binding or remodeling activities. These activities depend on a number of conserved sequence motifs that are more closely related within one family than between families. Enzymes grouped into the same family often share a number of additional domains that support and modulate the activities of the RecA domains. Yet further N-terminal and C-terminal regions or insertions are variable and specific for particular family members and can mediate interactions with substrates and cofactors or sub-cellular localization. We study DExH-box enzymes involved in pre-mRNA splicing and bacterial DExH-box enzymes. While the latter group of proteins has not been investigated extensively, they are thought to play important roles as co- or post-transcriptional gene regulators that facilitate the adaptation of bacteria to changing environments and stress conditions.
Recent publications
Pietrzyk-Brzezinska AJ, Absmeier E, Klauck E, Wen Y, Antelmann H, Wahl MC (2018) Crystal structure of the Escherichia coli DExH-box helicase HrpB. Structure 26, 1462-1473. (PubMed)
Absmeier E, Becke C, Wollenhaupt J, Santos KF, Wahl MC (2017) Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle 16, 100-112. (PubMed)
Henning LM, Santos KF, Sticht J, Jehle S, Lee CT, Wittwer M, Urlaub H, Stelzl U, Wahl MC, Freund C (2017) A new role for FBP21 as regulator of Brr2 helicase activity, Nucleic Acids Res, 13, 7922-7937. (PubMed)
Theuser M, Höbartner C, Wahl MC, Santos KF (2016) Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci USA 113, 7798-7803. (PubMed)
Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF, Wahl MC (2015) The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev 29, 2576-2587. (PubMed)