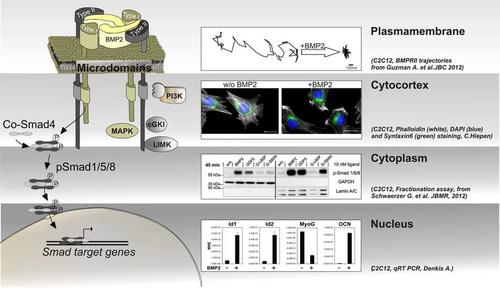
Subcellular compartmentalization of BMP signal transduction
Diversity in BMP signal transduction is achieved by activation of Smad and non‑Smad pathways including their mode of action in different subcellular compartments.
The Smad signalling cascade originates from pre-formed BMPRI and BMPRII receptor complexes and is fine tuned by and crosstalks to other pathways in the cytoplasm to finally act in the nucleus by activation of Smad dependent target genes. Non-Smad signalling, such as activation of MAPK, requires the formation of BMP-induced receptor complexes and may also act in different subcellular compartments, dependent on the cell context.
With respect to subcellular compartmentalization of BMP signalling, we therefore focus on:
- the plasma membrane, in particular the dynamics of BMP receptors within distinct membrane microdomains and the role of membrane scaffolding proteins in segregating and facilitating specific pathways. Here, we establish single particle tracking microscopy (SPT) of the receptors to follow the differential lateral mobility of BMPRI and BMPRII under defined stimulation conditions. Our current aim is to understand mechanisms of confinement, the establishment of diffusion barriers but also active transport mechanisms of BMP receptors to and from the plasma membrane and how this integrates into BMP signalling intensity, specificity and duration.
- the cytocortex which represents the most proximal cytoplasmic region below the plasma membrane of polarized cells. Here BMPs induce heavy and quick cytoskeletal rearrangements but also recruitment/tethering of scaffolding proteins and the production/enrichment of distinct lipids. Application of Mass Spectrometry revealed novel BMP receptor interacting proteins that produce lipids or show a lipid dependency but also act as cytoskeletal regulators. To investigate novel BMP dependent molecular mechanisms in respect to planar cell polarity, cell migration and endo-/exo-cytosis we use lightoptical techniques such as confocal-, super-resolution-, and TIRF-microscopy.
- the cytoplasm as well as cytoplasmic compartments such as endocytic or exocytic vesicles. Here we aim to understand where and when BMP pathway proteins are transported and fine tuned and how they integrate and crosstalk to other pathways such as Wnt, Notch or Hippo cascades. As shown by others, this crosstalk appears also on the level of Smad linker phosphorylation leading to degradation of Smads. We use proteomics and protein-protein biochemistry to identify novel regulatory frameworks as well as life cell imaging and biosensors to visualize translocation events in real time.
- the nucleus including the nuclear envelope. Entrance of Smads into the nucleus including assembly of transcriptional complexes pre or post entrance may fine tune the transcriptional regulation of Smad dependent target genes. In that respect we are interested in kinetics of Smad nuclear translocation but also into identifying novel Smad transcriptional complexes involved in osteogenic, myogenic and endothelial differentiation.