Forschung
Understanding Neural Circuits
Across animal brains, the interactions between many neurons control a variety of behavioral responses. How does that happen? In order to understand how the brain works on a cellular level, we use a combination of very different approaches: behavior experiments, neuroanatomy, and (Opto-)physiology.
Neural Circuits and behavior
We would like to understand the role of identified neuronal cell types while specific behaviors are being executed. We use the molecular genetic tools available in the fruit fly Drosophila melanogaster to characterize these cell types, as well as to perturb their activity in the awake, behaving animals. Following this strategy, we can switch specific neuronal populations ‘on’ and ‘off’ with high special and temporal precision. We then assess the consequences of these manipulations on the behavioral performance of the animal. Of course, the behavioral repertoire of a fly is very rich – just try to catch one! For historic reasons, we focus on responses to visual stimuli, which have been studied in flies for many decades, resulting in elegant computational models. Flies can see (and learn) shapes, they can perceive moving edges of different intensities (moving or looming objects), but they can also distinguish and compare different wavelengths (color vision), or perceive the e-vector of the incoming light (polarization vision). Together, these stimuli can be used by the animal to improve its navigational skills, when maneuvering through the environment. In order to study these behavioral responses quantitatively, we use classic behavior assays like a fly walking on an air-suspended ball, or flying in a virtual flight arena (a ‘flight simulator’), while experiencing different visual stimuli. Hence, following this strategy, quantitative changes in the behavioral response can be linked to a neuronal substrate.
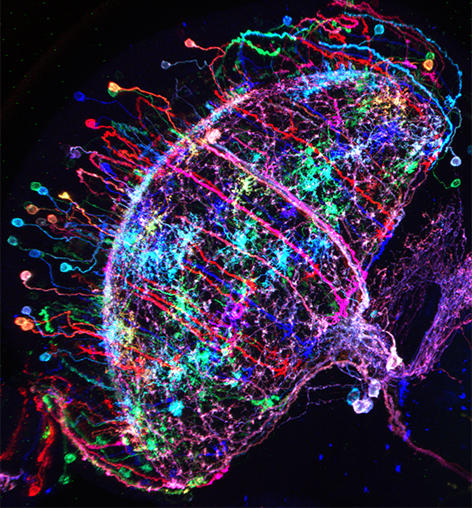
Figure 1: ‘Brainbow’-like labeling of individual Drosophila neurons in multiple colors
Neural Circuits and connectivity
Additional pieces of information can be helpful towards understanding what a neural circuit is doing. For instance, which cell types are we looking at, and
how are they synaptically connected? Thankfully, we can use a great arsenal of tools for answering these very questions. For instance, two large public libraries of transgenic GAL4 (and in some cases, LexA) driver lines give us reproducible access to most, if not all cell types in the fly brain (the brain may be small, but we are talking 100,000+ cells!). In both cases, intronic enhancer sequences from the fly genome were used in an unbiased way, to label small groups of neurons (https://www.janelia.org/open-science/gal4-fly-lines and http://stockcenter.vdrc.at/control/library_vt). Using stochastic labeling techniques like ‘brainbow’ or ‘multiple color flip-out’, individual cells can be visualized and characterized, resulting in the unambiguous identification of the studied cell types. Their input and output regions can be labeled with fluorescent proteins localized either to synaptic or dendritic compartments. But what about identified synaptic connections? The problem is that synapses are very small and cannot be seen using light microscopy, at least not without using certain tricks. One elegant method uses the reconstitution of green-fluorescent proteins across synaptic partners (GRASP). If two cells form very intimate contacts, for instance at synapses, the two complementary halves of GFP they both express can reconstitute the full fluorescent protein only there, thereby specifically marking the synapses between these two identified cell types through a fluorescent signal. But that’s not all: More recently, the entire fly brain is being reconstructed at electron-microscopic resolution, meaning that in the not-so-distant future, all synaptic connections between our neurons of interest will be known! (see https://www.janelia.org/project-team/flyem).

Figure 2: A fly walking on an air-suspended ball
Neural Circuits and activity
For quite a while now, the entire nervous system of the namatode Caenorhabditis elegans has been reconstructed in 3D and at electron microscopic resolution, meaning all the synapses between all neurons are known for this species. Does that mean the end of nematode Neuroscience? No. But why? The reason is that neuronal circuits weren’t built by engineers with a particular purpose in mind. Instead, we need one more piece of information, besides connectivity and the neuron’s impact on the behavioral responses of the animal. This missing information is the pattern of excitation of the individual neurons, and how it is modulated by our (in our case: visual) stimulus. The study of neuronal activity has of course a very long and successful tradition. For most of this time, people have used different forms of electrophysiology to investigate cellular responses (for instance, the frequency of action potentials). However, since fly neurons are very small, it is not always possible to use electrophysiology. Instead, our field is using the advantages of Optophysiology. Using genetically encoded calcium sensors like GCaMP6, the activity of a neuron can be visualized indirectly, using a 2-photon microscope which allows visualization in vivo. Using this technique, the responses of neurons deep within the brain can be visualized, by imaging through a small window which has been cut into the back of the fly’s head. With this tool in hand one can now interrogate the structure and function of neural circuits at multiple levels, covering everything from anatomy, behavior and activity.
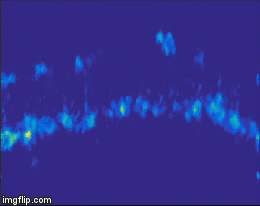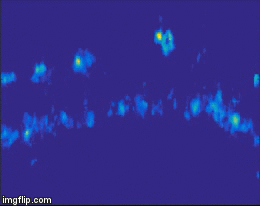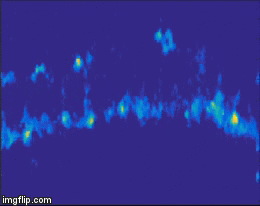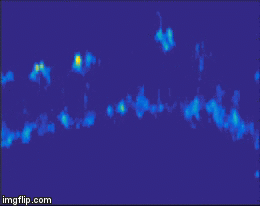
Figure 3: Calcium signals in the fly brain
